Host Protein General Information
| Protein Name |
Phospholipase C-II
|
Gene Name |
PLCG1
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Host Species |
Homo sapiens
|
Uniprot Entry Name |
PLCG1_HUMAN
|
||||||
| EC Number |
3.1.4.11
|
||||||||
| Subcellular Location |
Cell projection; lamellipodium
|
||||||||
| External Link | |||||||||
| NCBI Gene ID | |||||||||
| Uniprot ID | |||||||||
| Ensembl ID | |||||||||
| HGNC ID | |||||||||
| Related KEGG Pathway | |||||||||
| Chemokine signaling pathway | hsa04062 |
Pathway Map 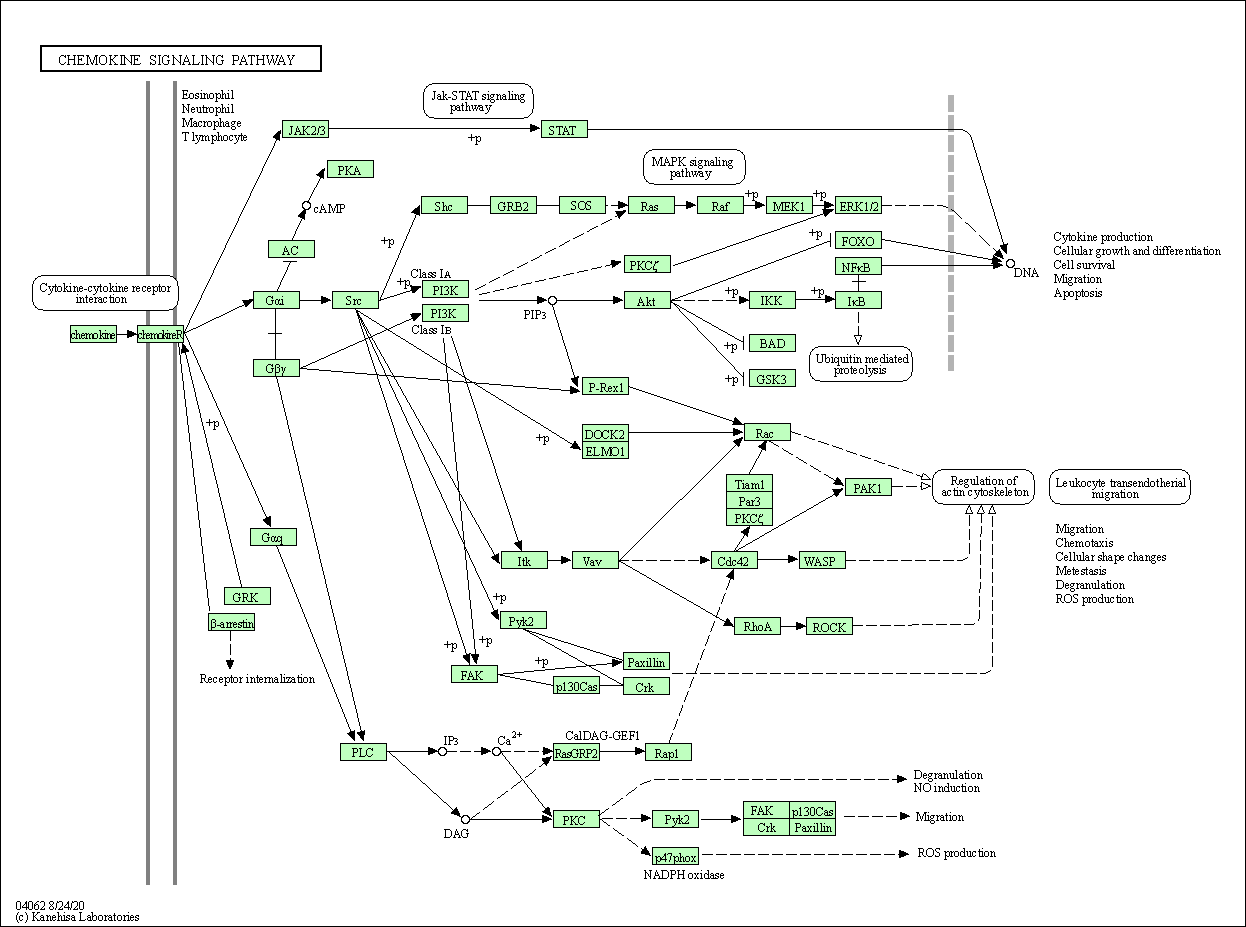
|
|||||||
| Neutrophil extracellular trap formation | hsa04613 |
Pathway Map 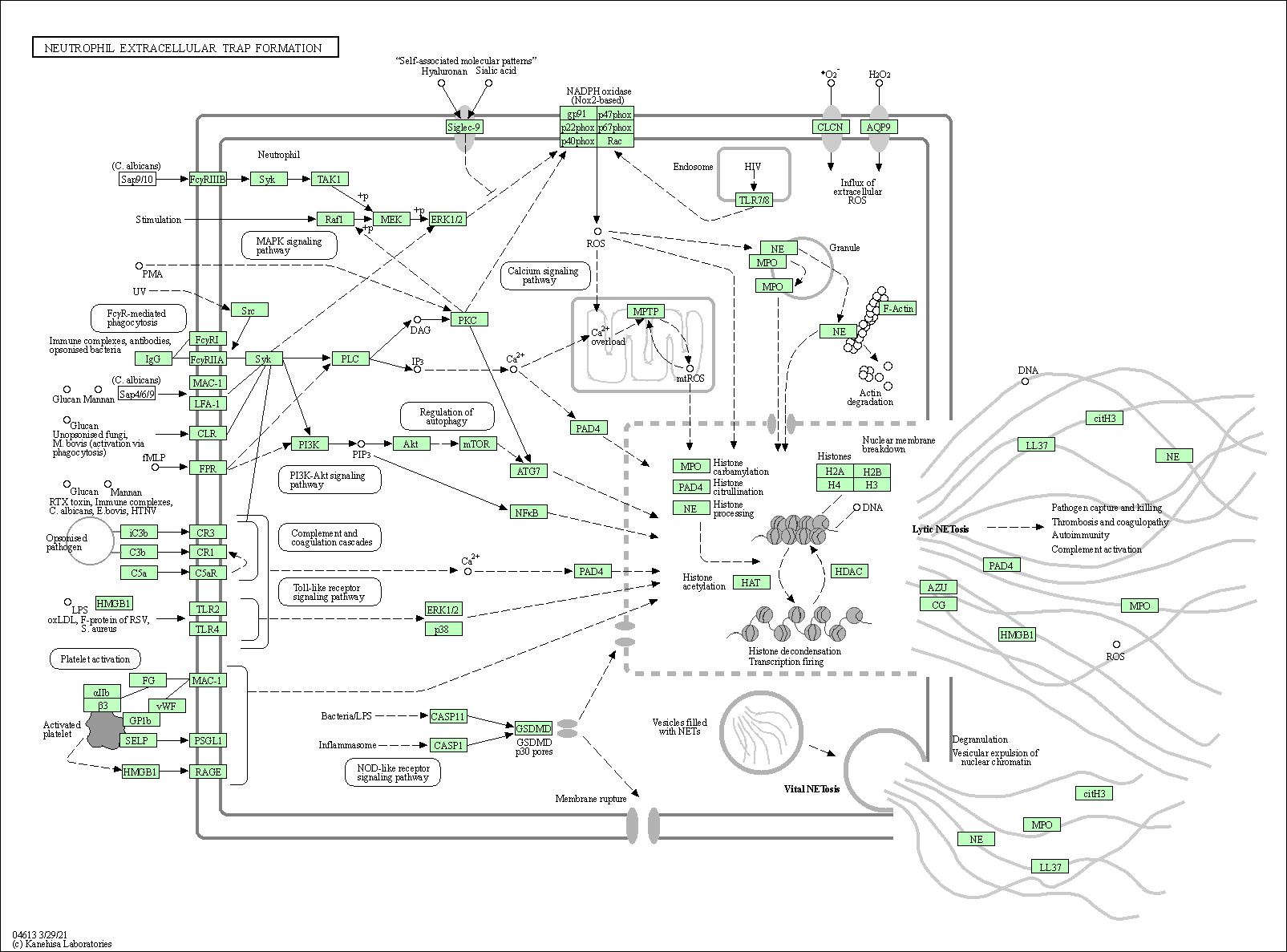
|
|||||||
| Natural killer cell mediated cytotoxicity | hsa04650 |
Pathway Map 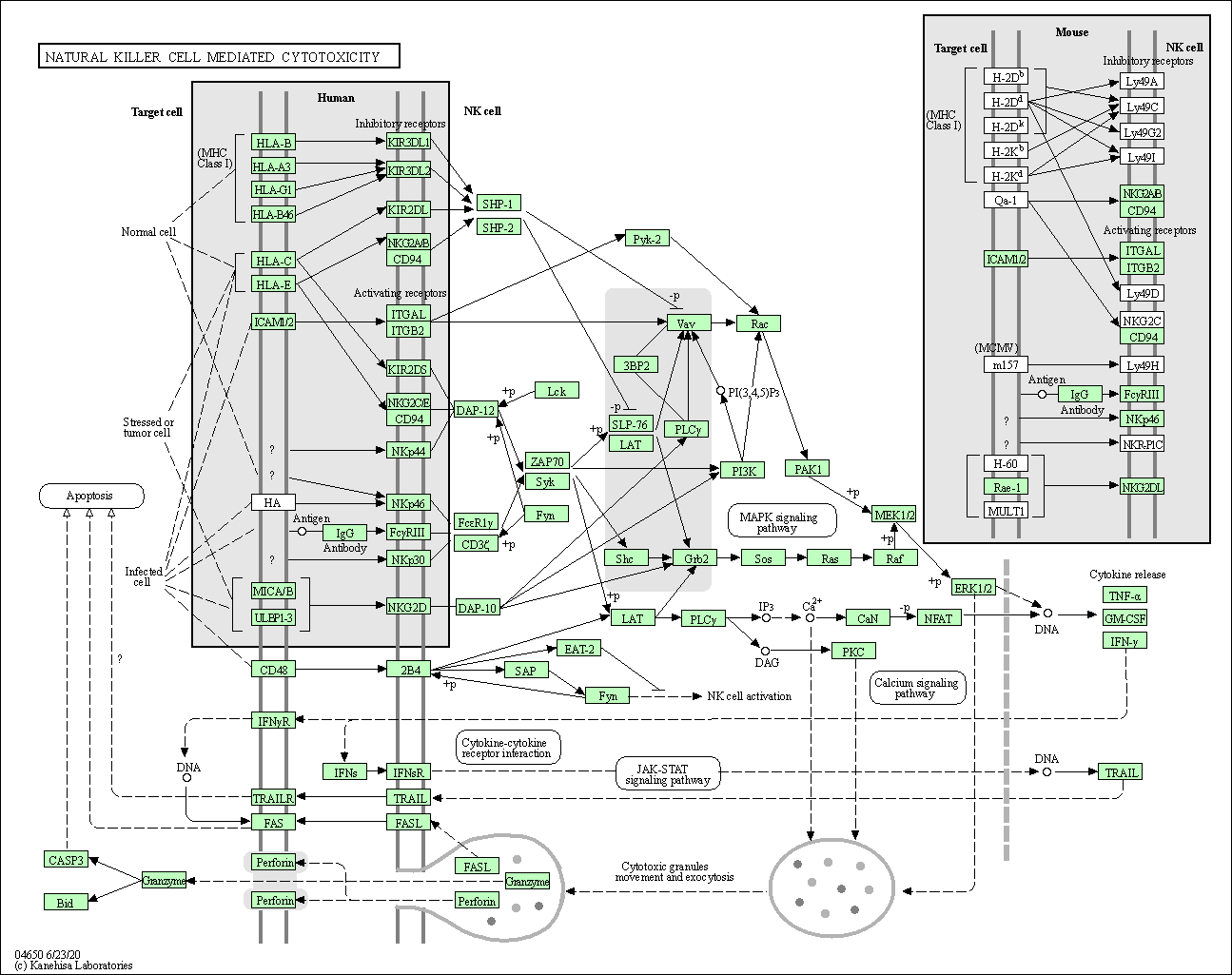
|
|||||||
| Th1 and Th2 cell differentiation | hsa04658 |
Pathway Map 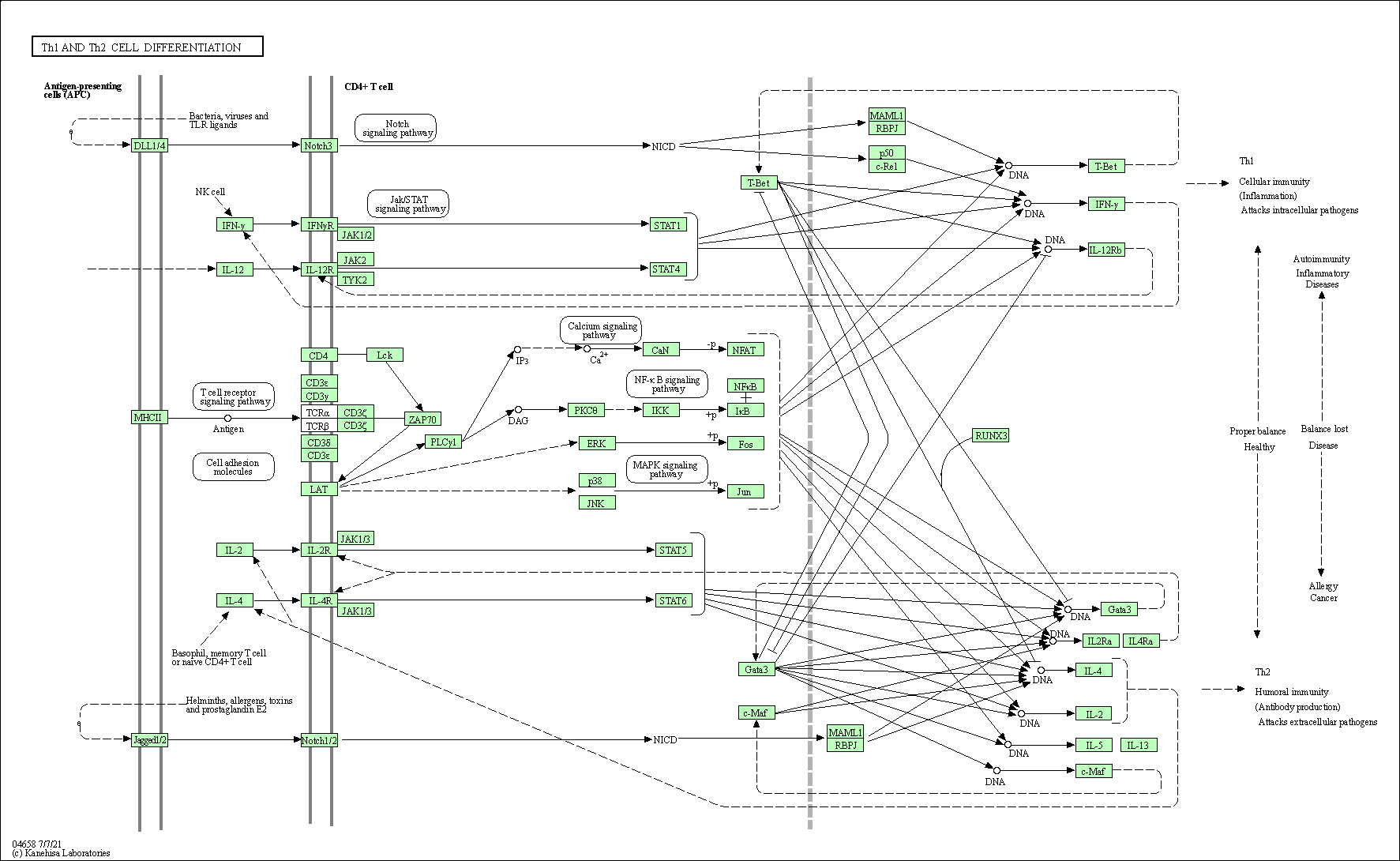
|
|||||||
| Th17 cell differentiation | hsa04659 |
Pathway Map 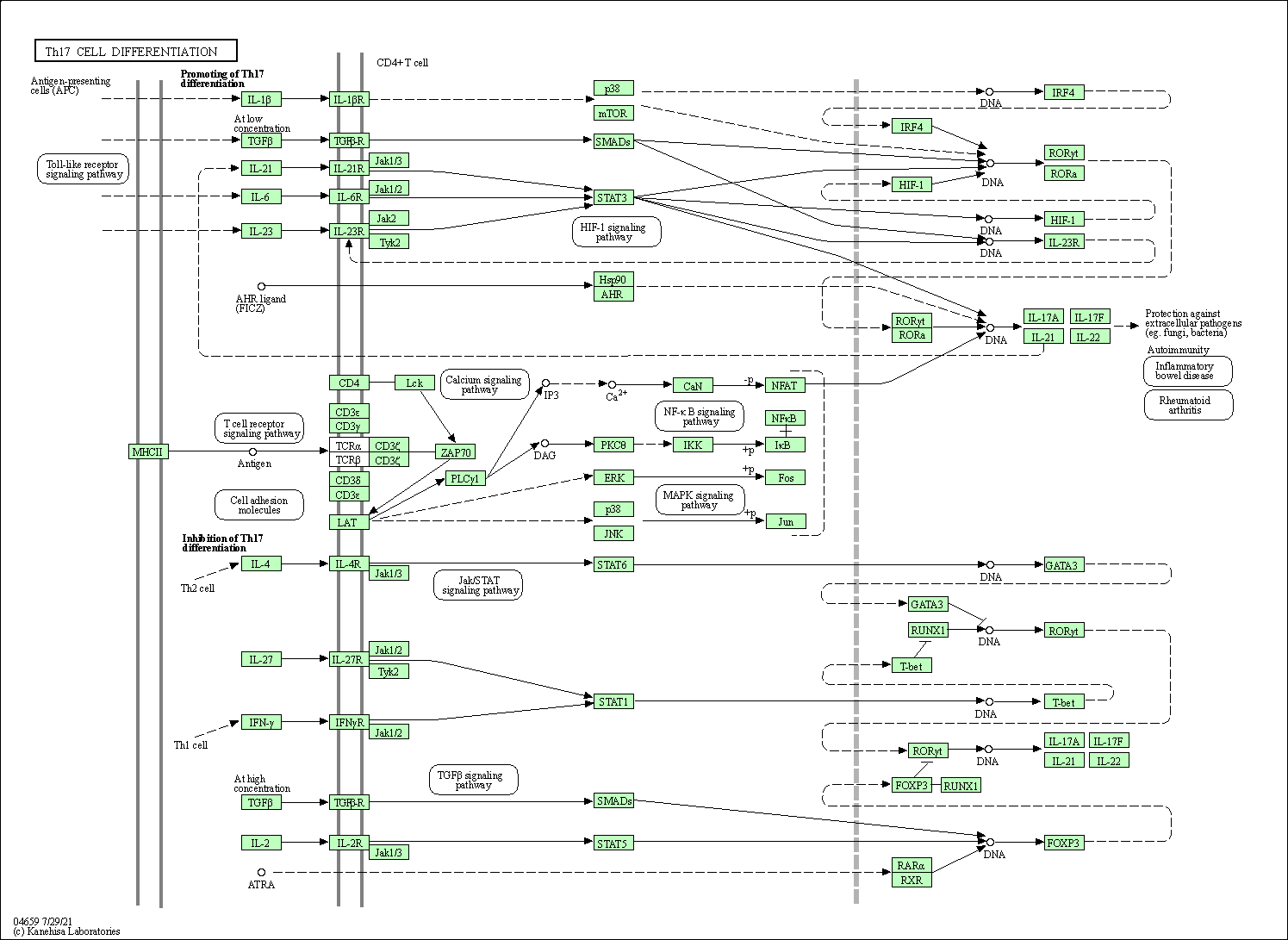
|
|||||||
| T cell receptor signaling pathway | hsa04660 |
Pathway Map 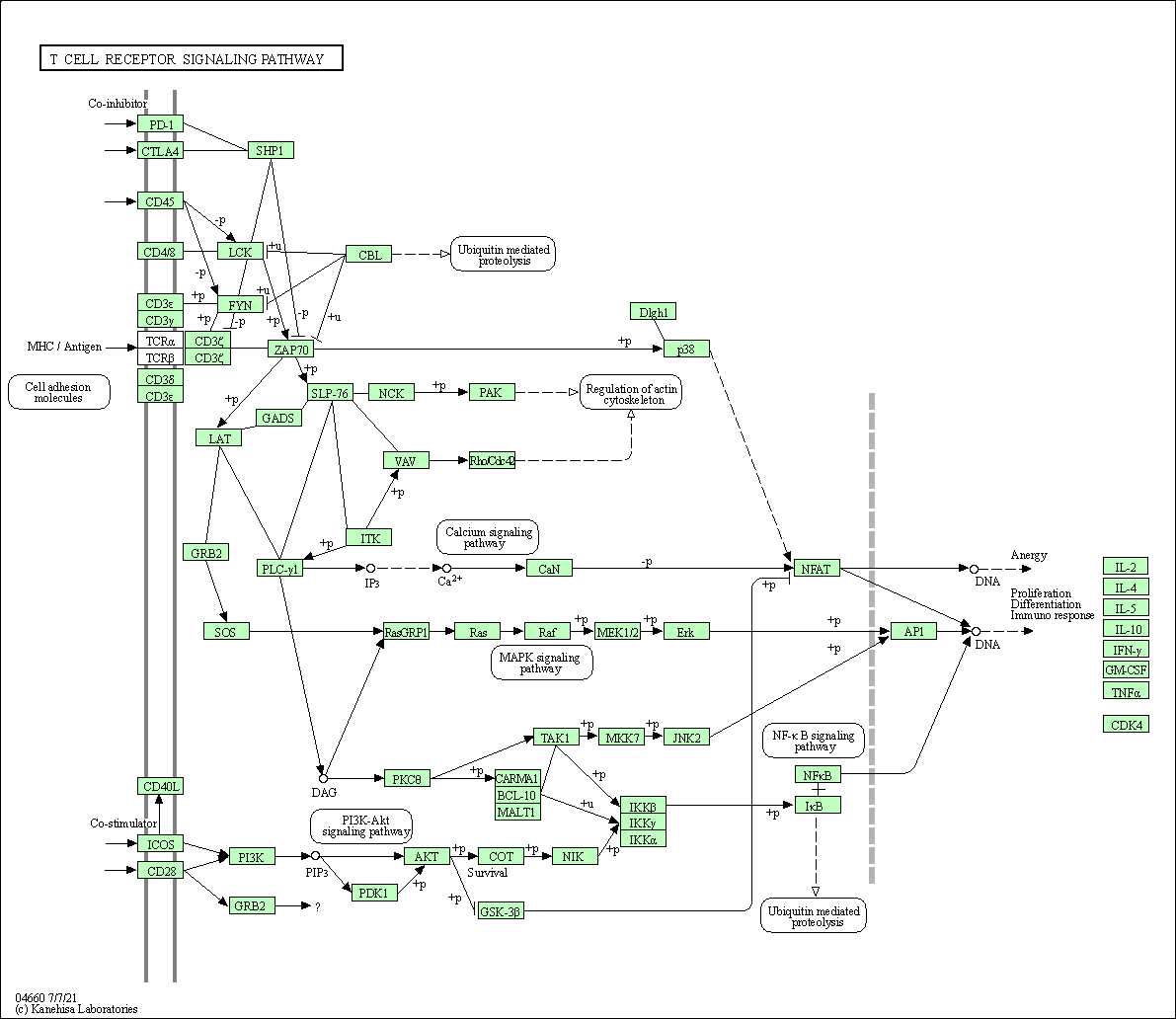
|
|||||||
| Fc epsilon RI signaling pathway | hsa04664 |
Pathway Map 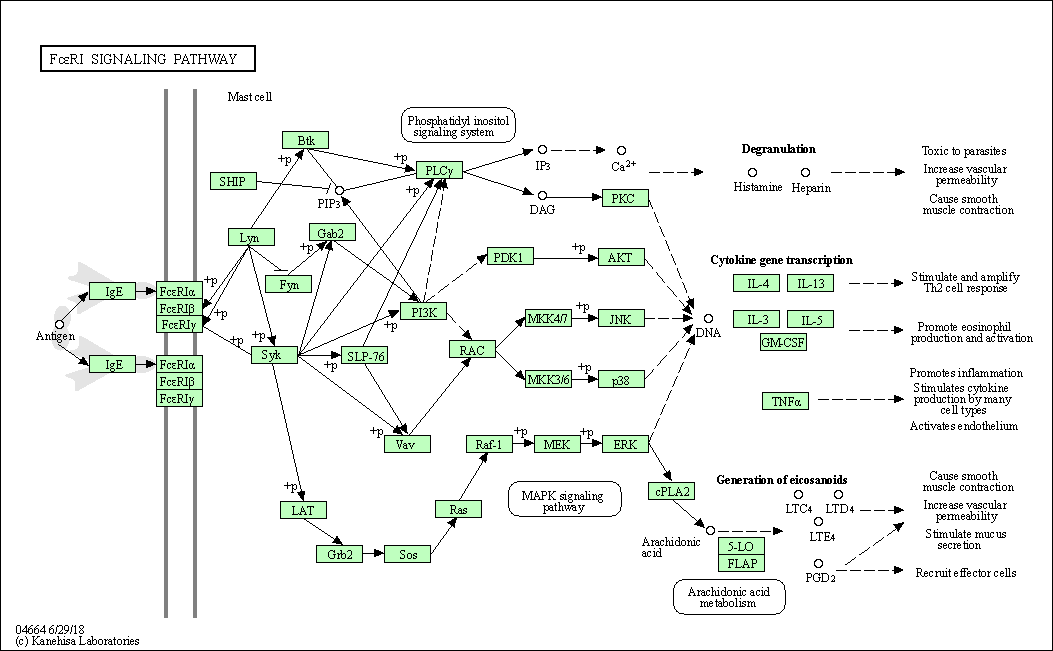
|
|||||||
| Fc gamma R-mediated phagocytosis | hsa04666 |
Pathway Map 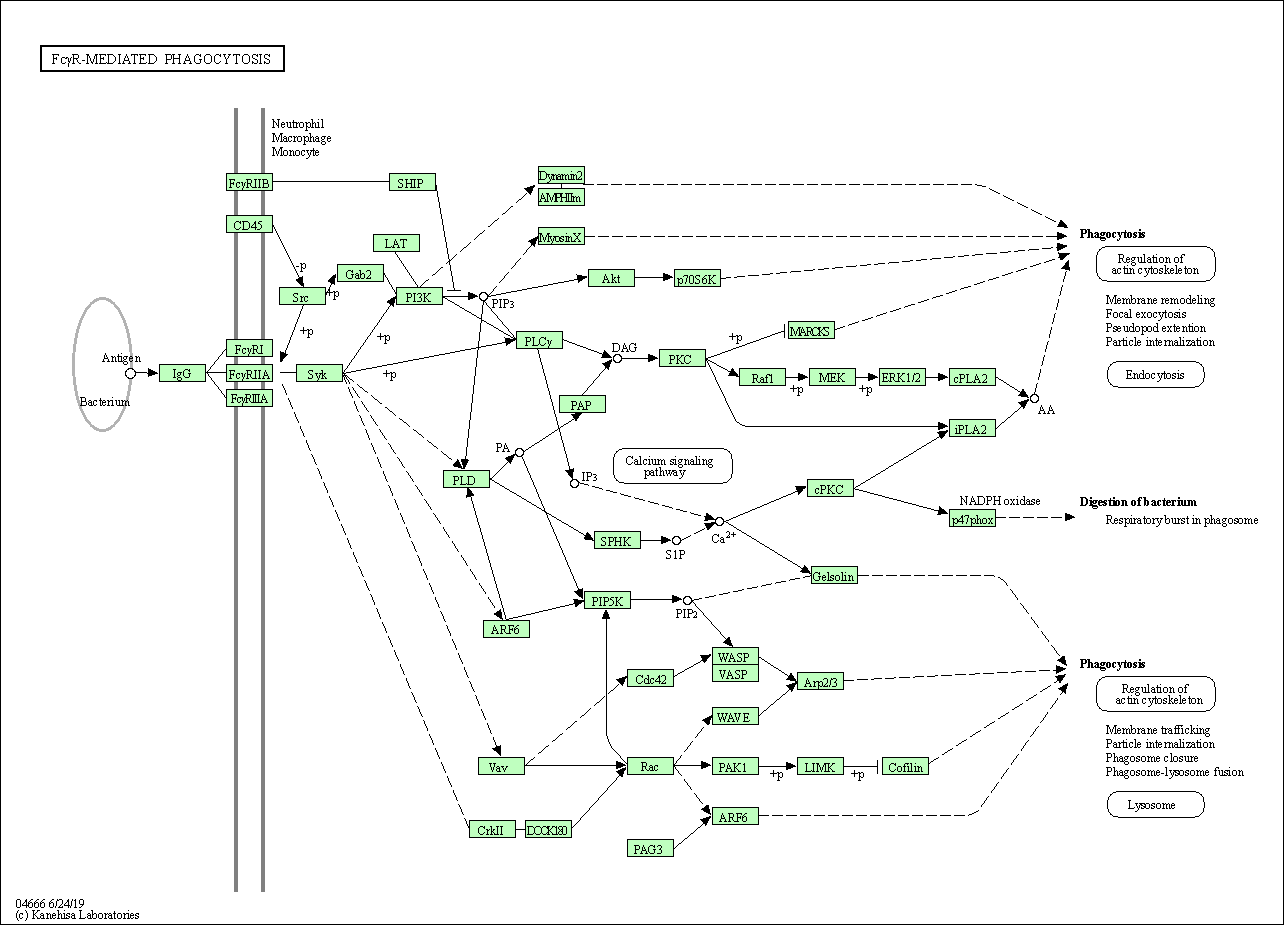
|
|||||||
| Leukocyte transendothelial migration | hsa04670 |
Pathway Map 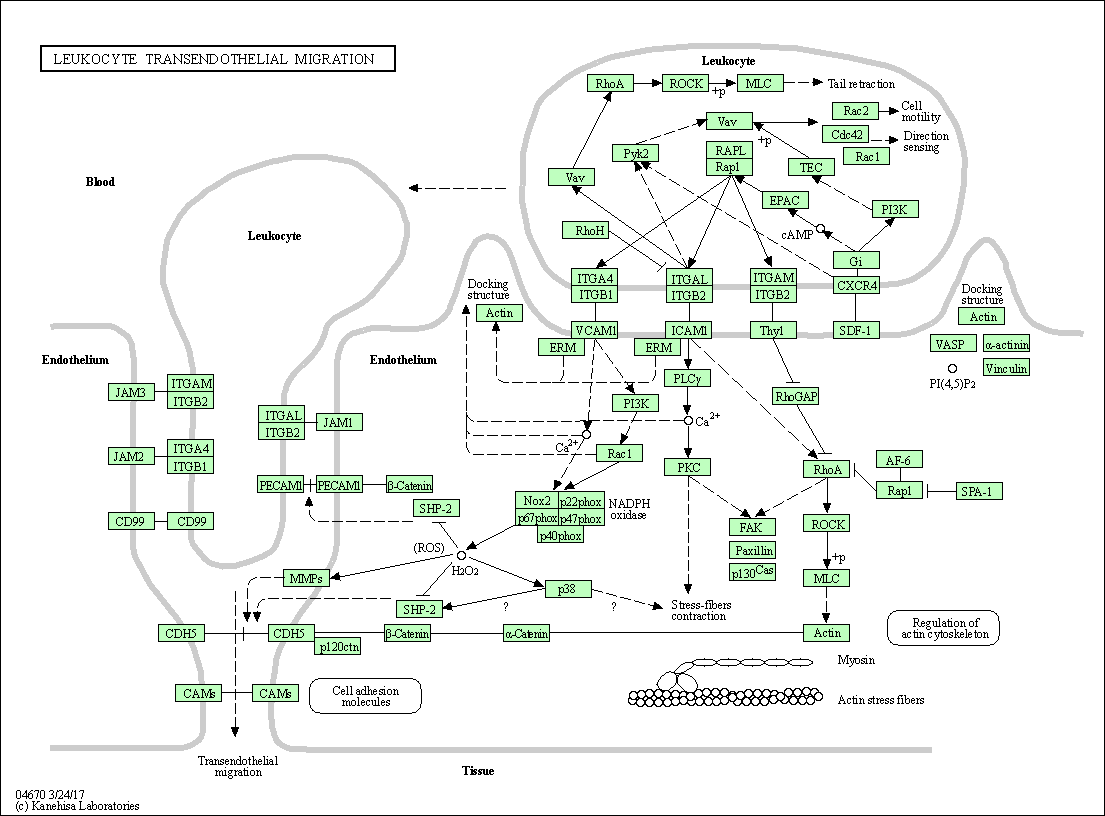
|
|||||||
| Vibrio cholerae infection | hsa05110 |
Pathway Map 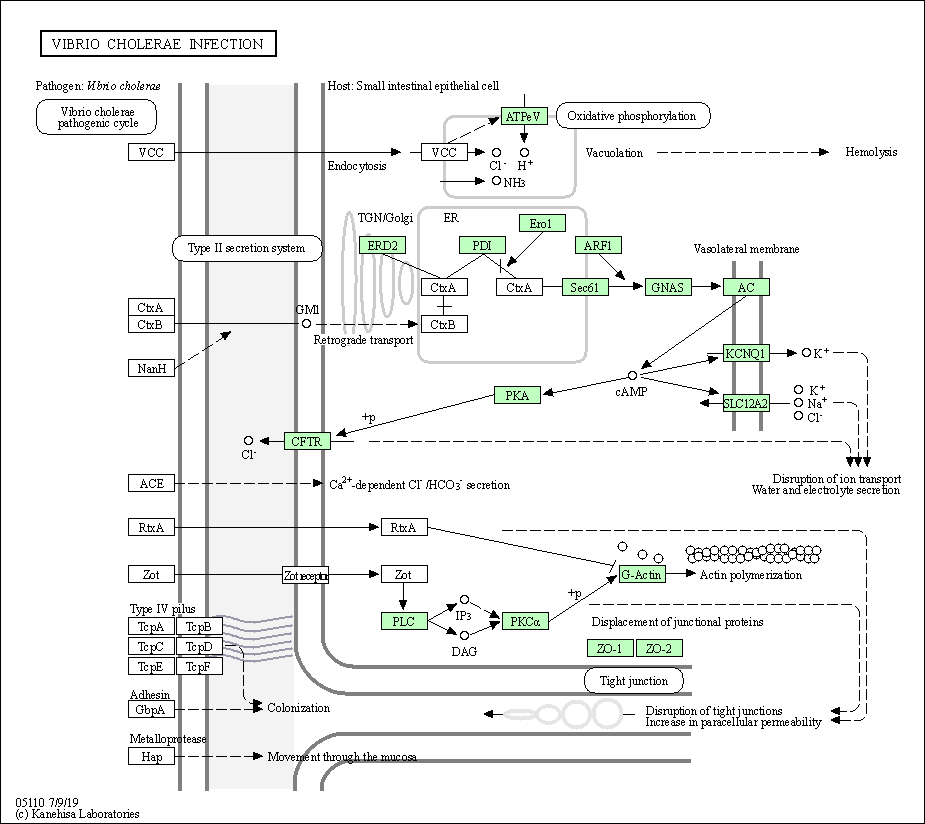
|
|||||||
| Epithelial cell signaling in Helicobacter pylori infection | hsa05120 |
Pathway Map 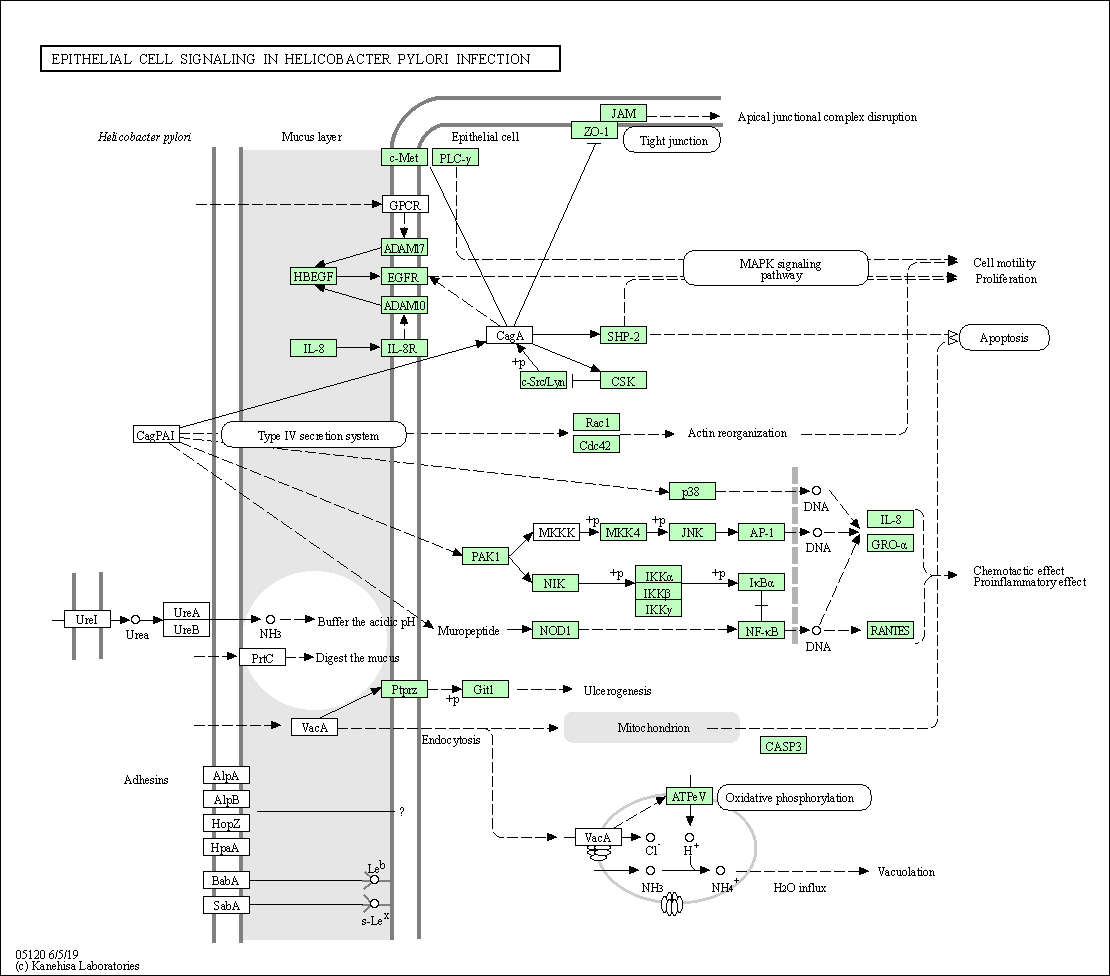
|
|||||||
| Shigellosis | hsa05131 |
Pathway Map 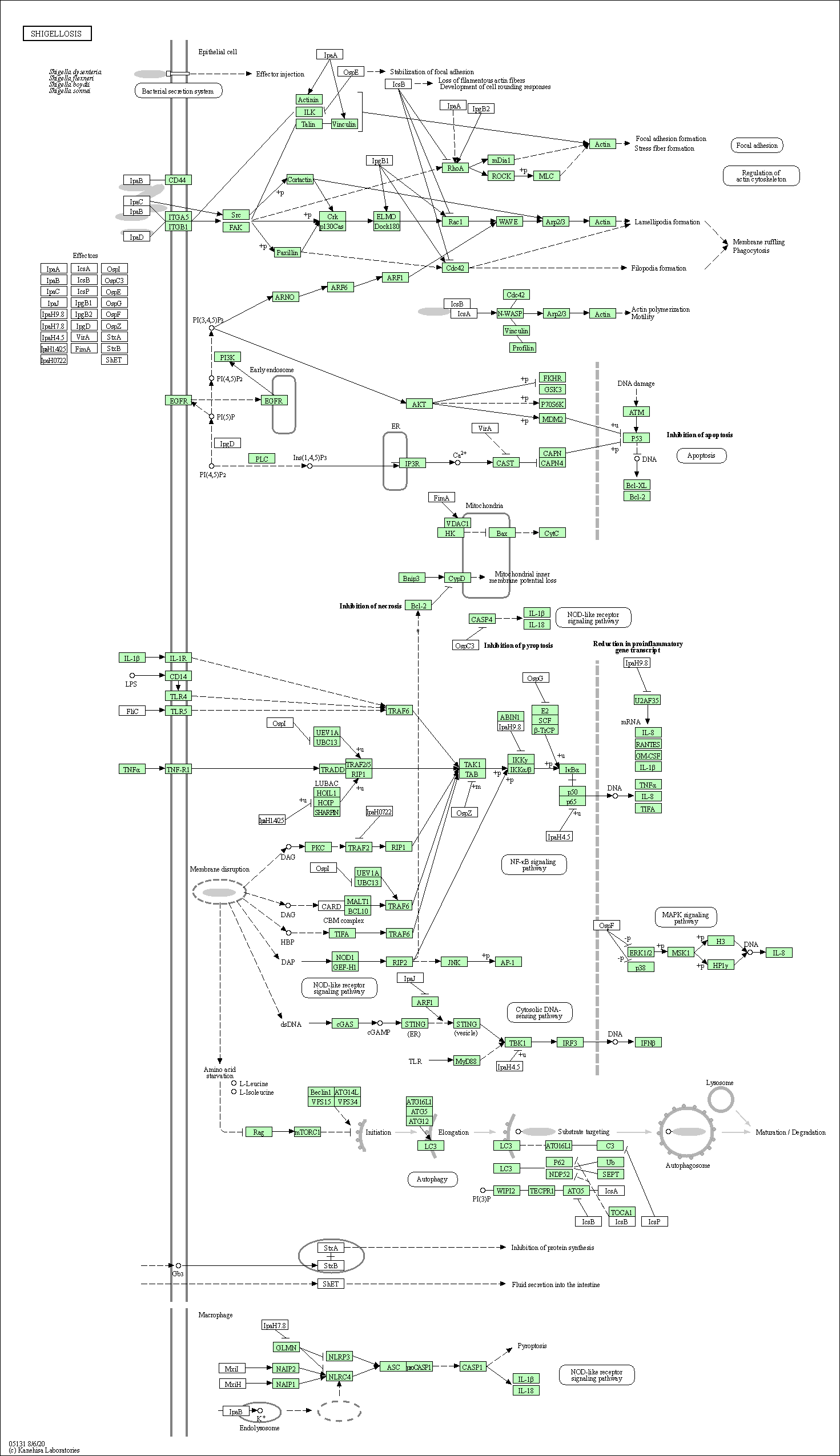
|
|||||||
| Yersinia infection | hsa05135 |
Pathway Map 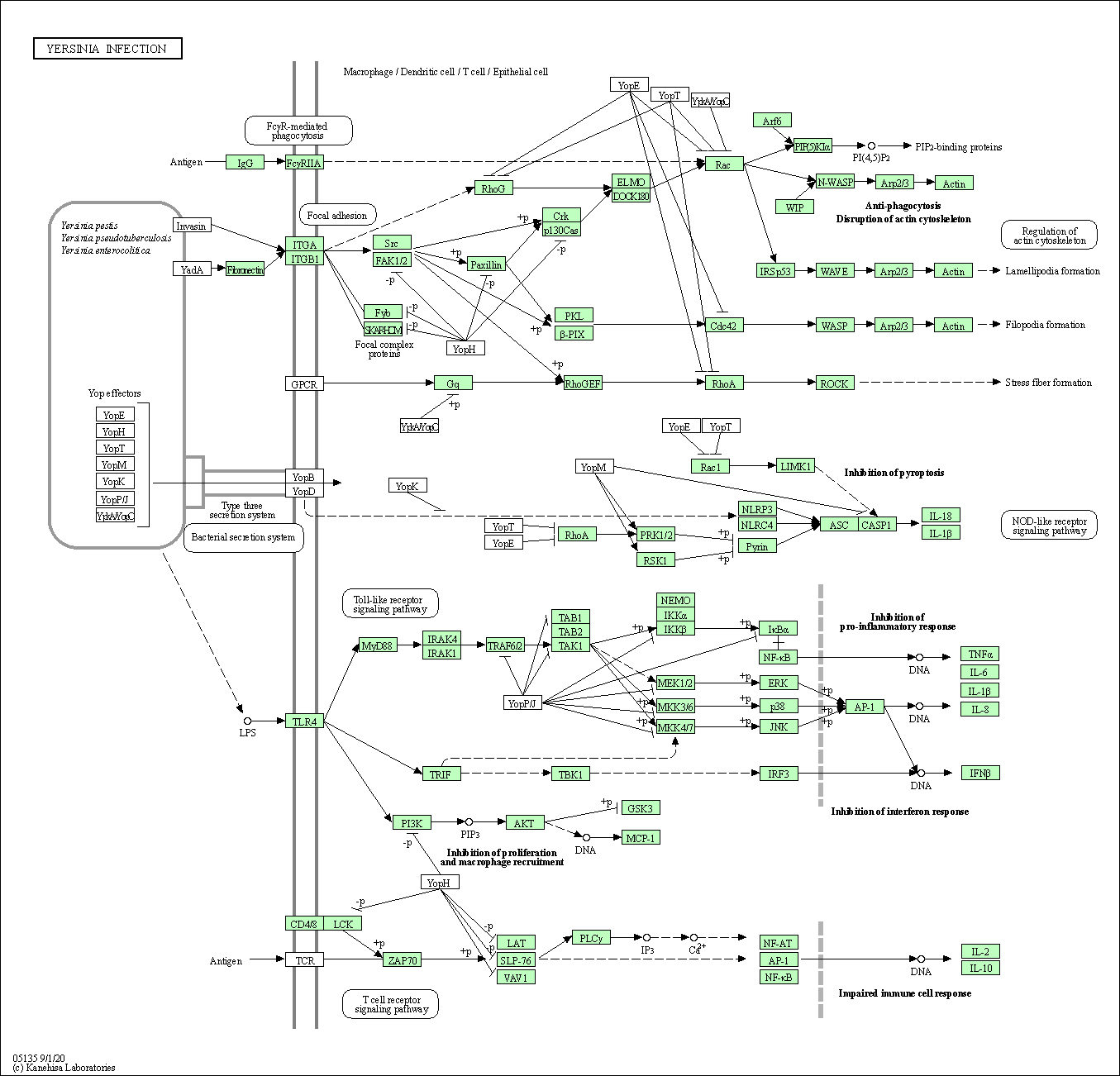
|
|||||||
| Kaposi sarcoma-associated herpesvirus infection | hsa05167 |
Pathway Map 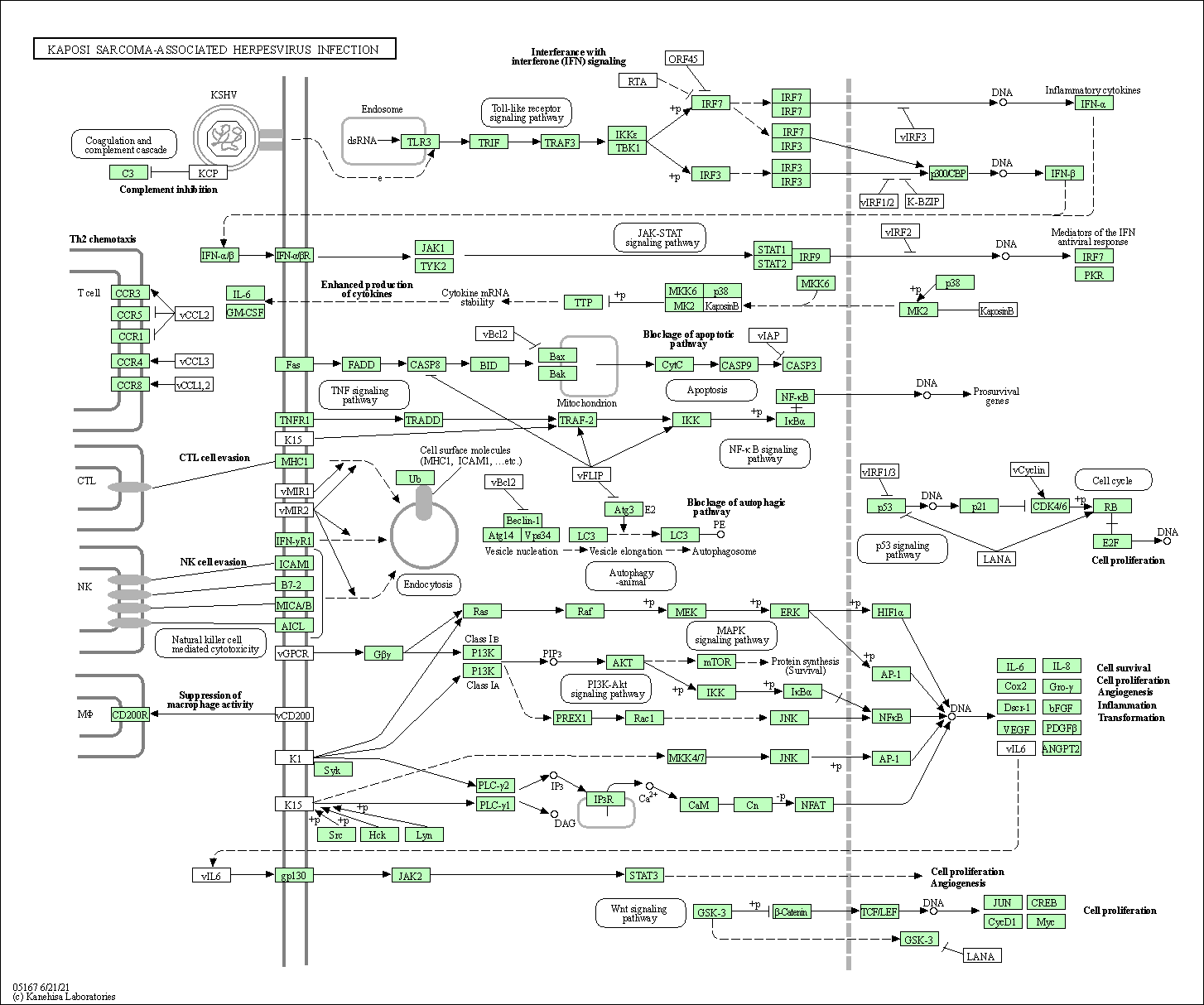
|
|||||||
| Human immunodeficiency virus 1 infection | hsa05170 |
Pathway Map 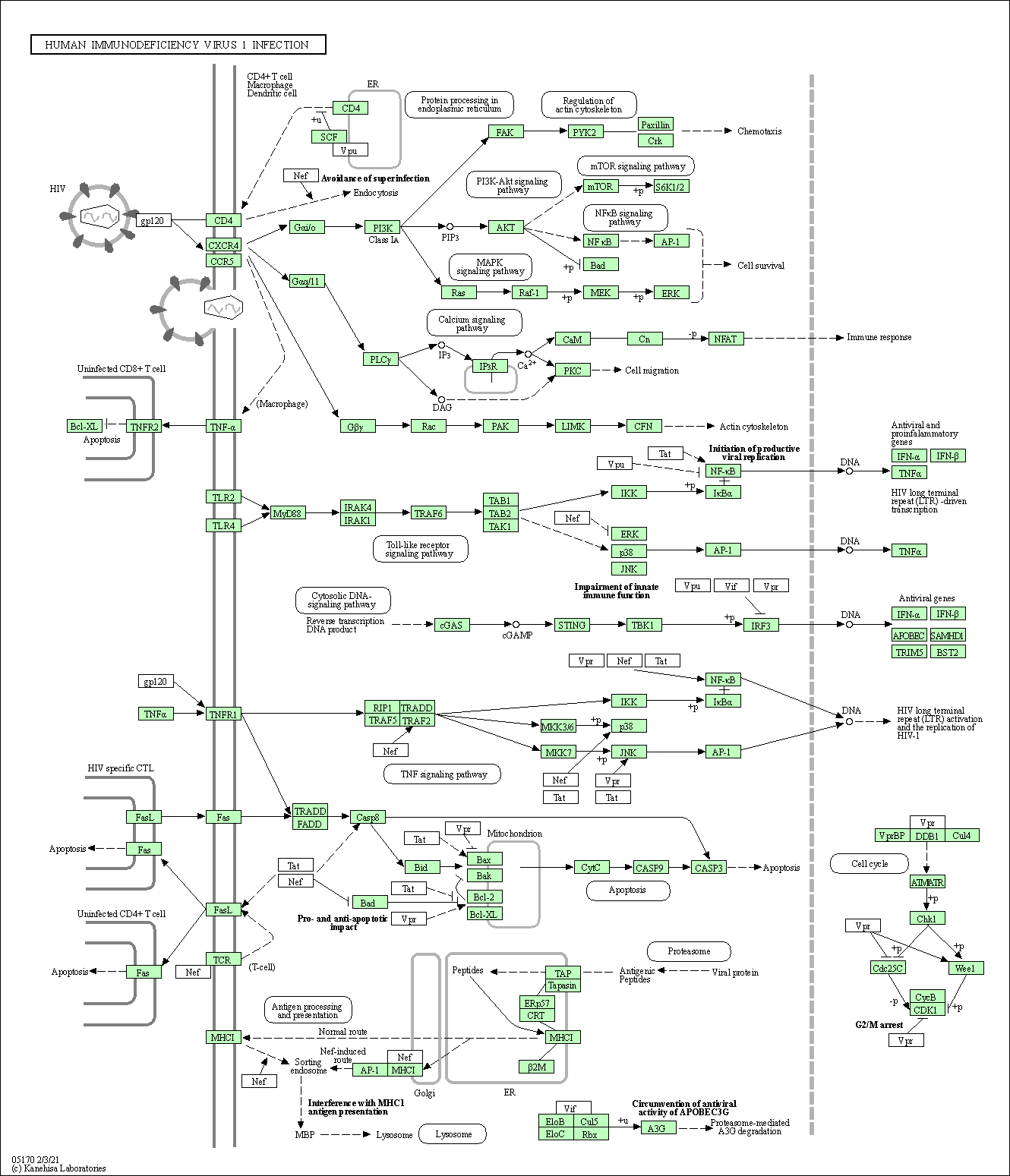
|
|||||||
| Coronavirus disease - COVID-19 | hsa05171 |
Pathway Map 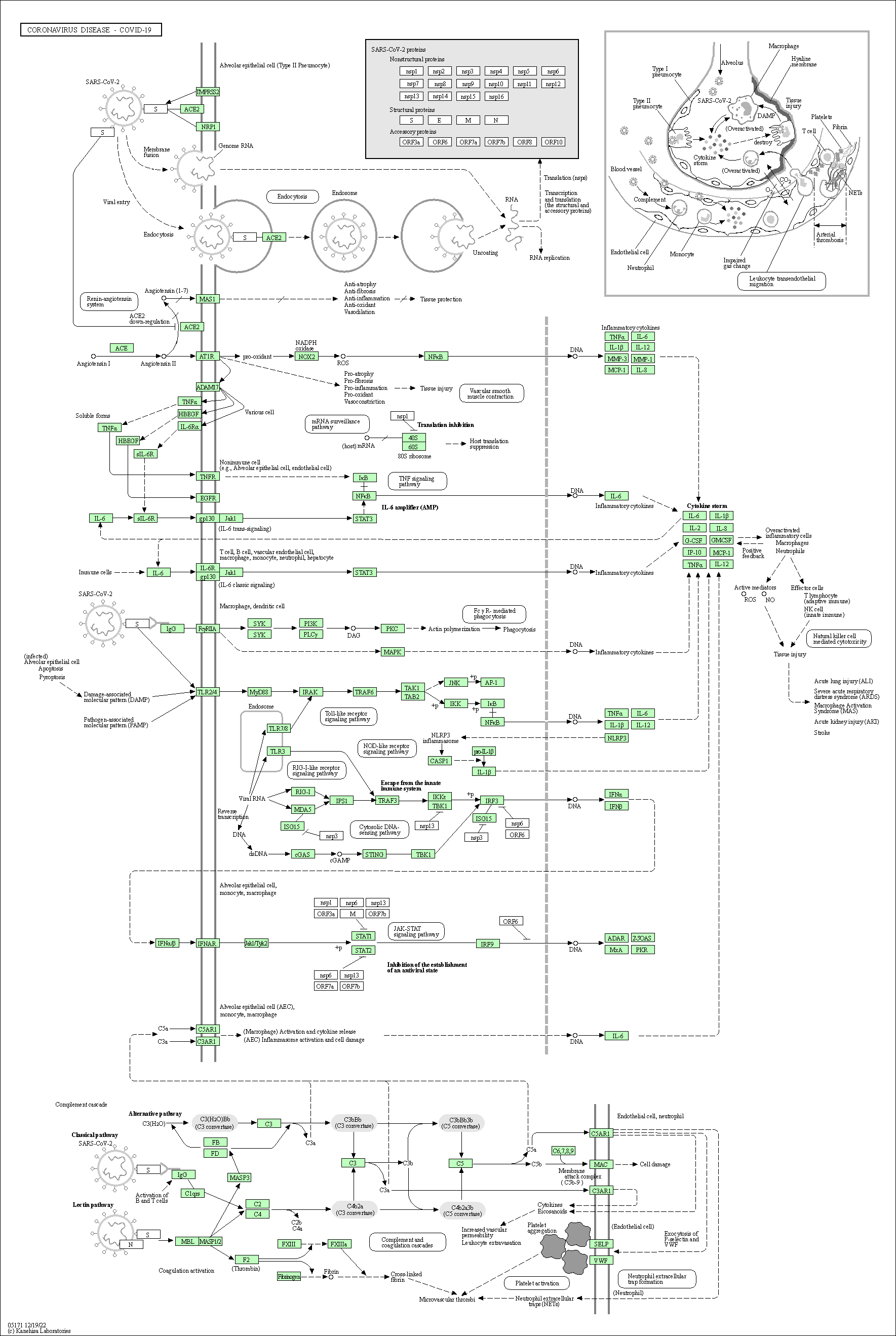
|
|||||||
| EGFR tyrosine kinase inhibitor resistance | hsa01521 |
Pathway Map 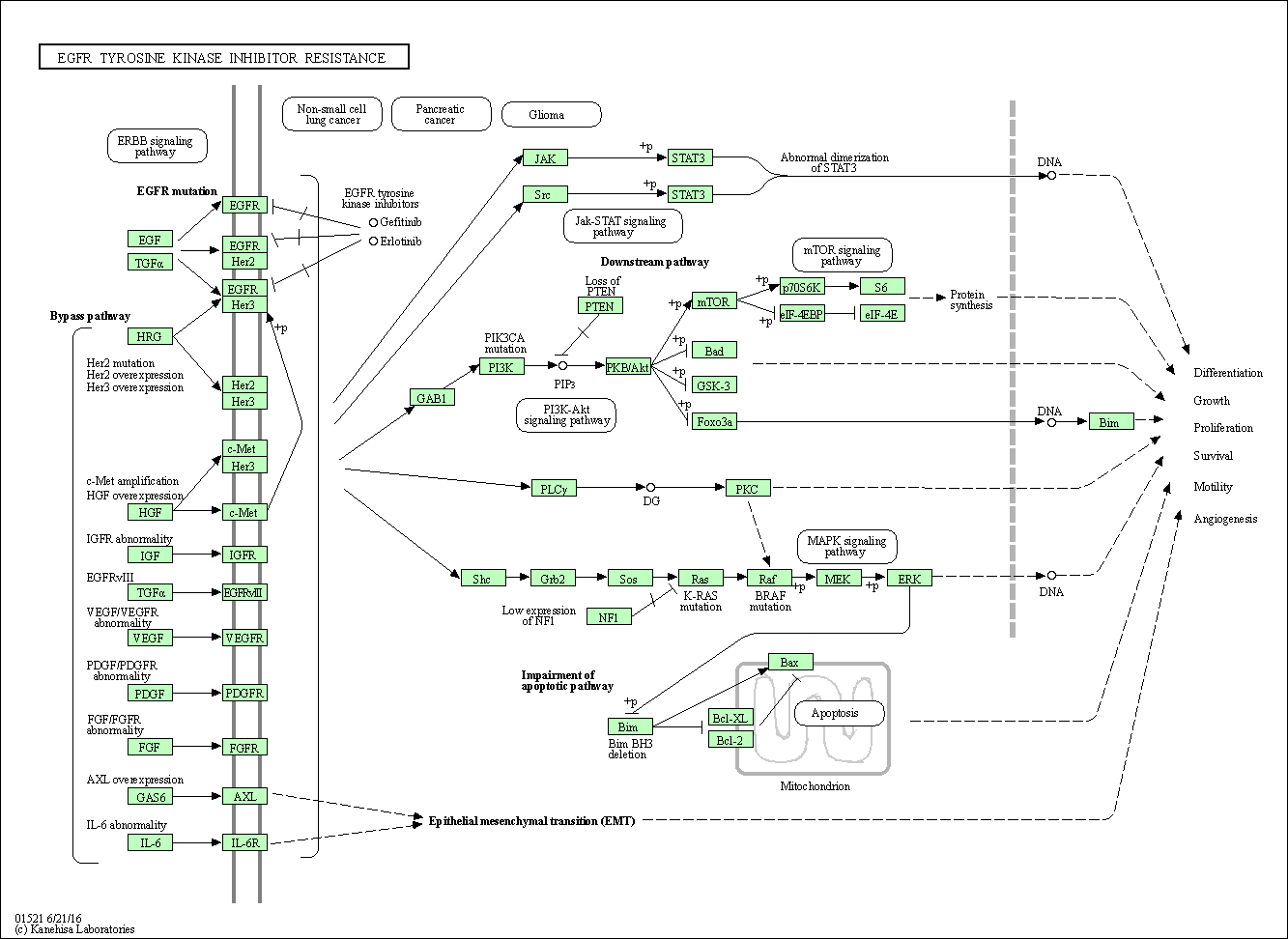
|
|||||||
| Pathways in cancer | hsa05200 |
Pathway Map 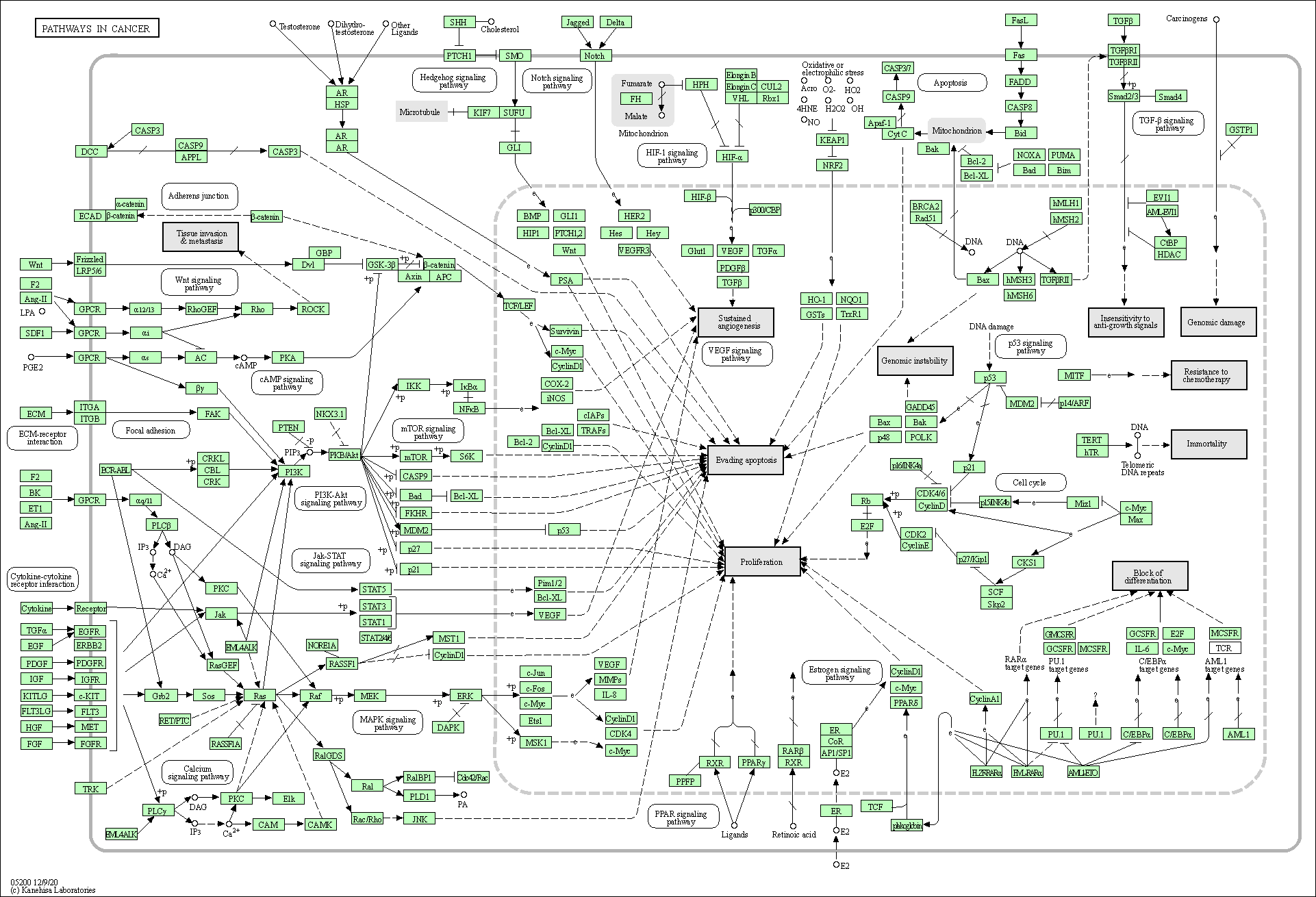
|
|||||||
| Proteoglycans in cancer | hsa05205 |
Pathway Map 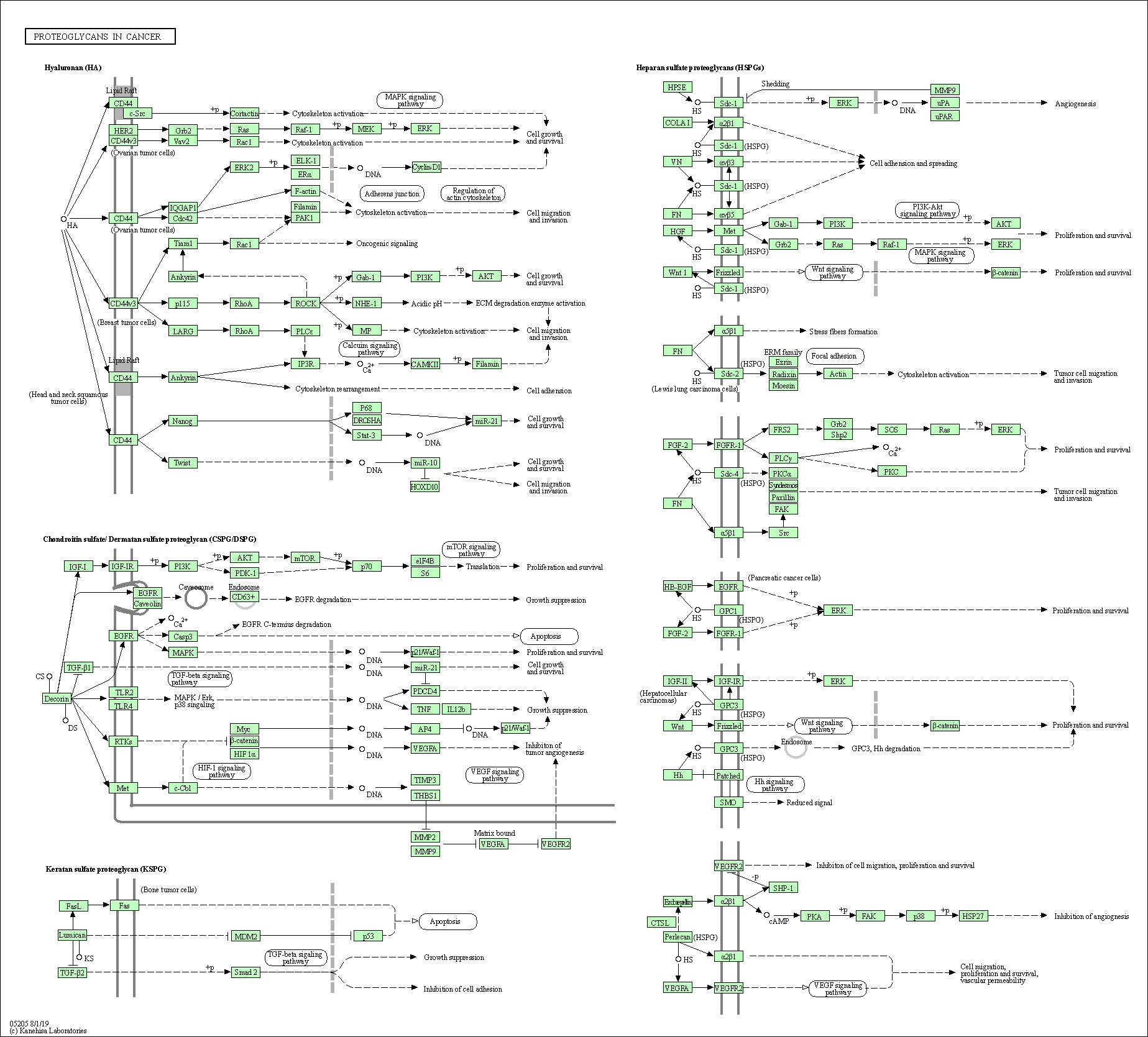
|
|||||||
| MicroRNAs in cancer | hsa05206 |
Pathway Map 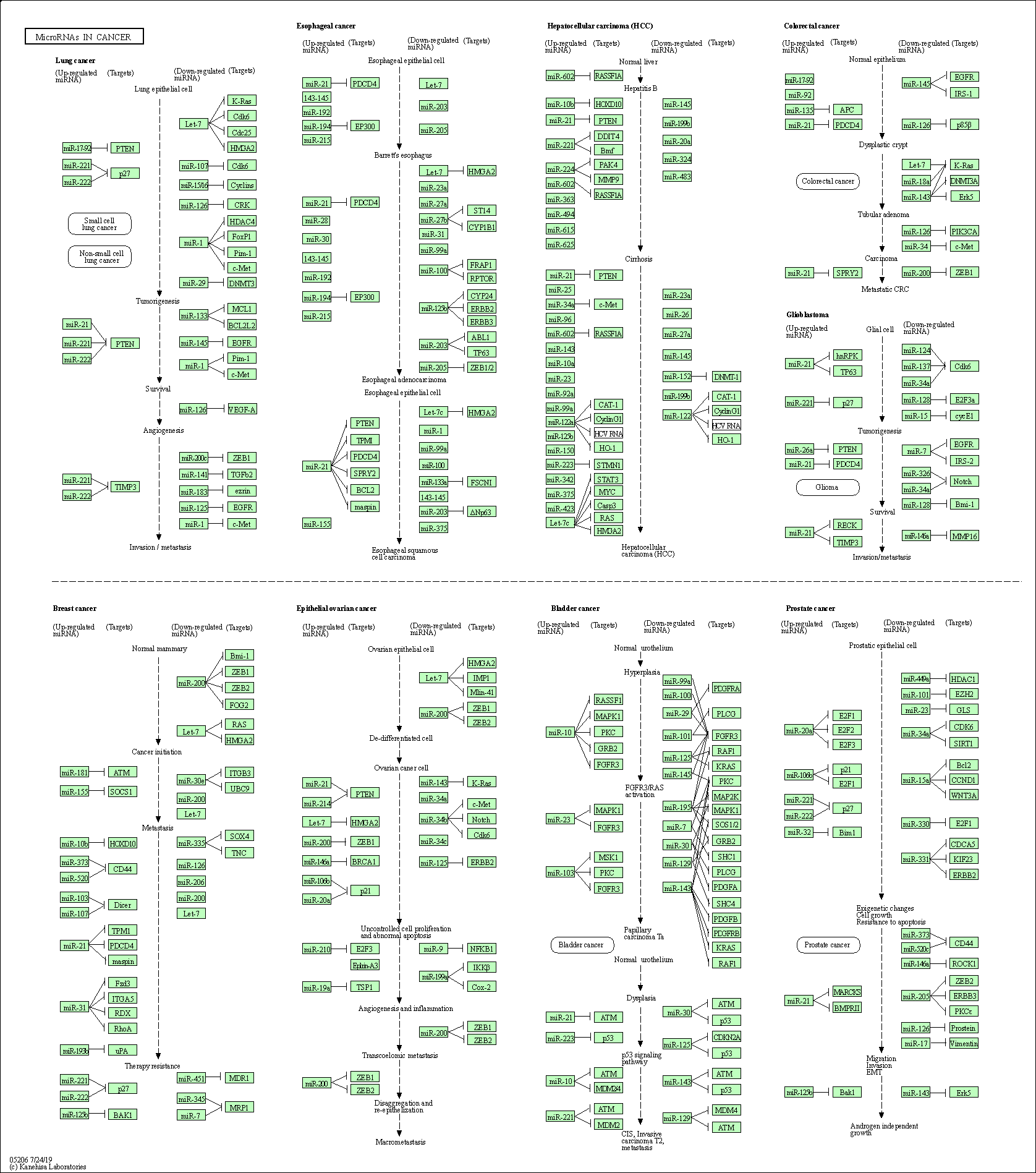
|
|||||||
| Choline metabolism in cancer | hsa05231 |
Pathway Map 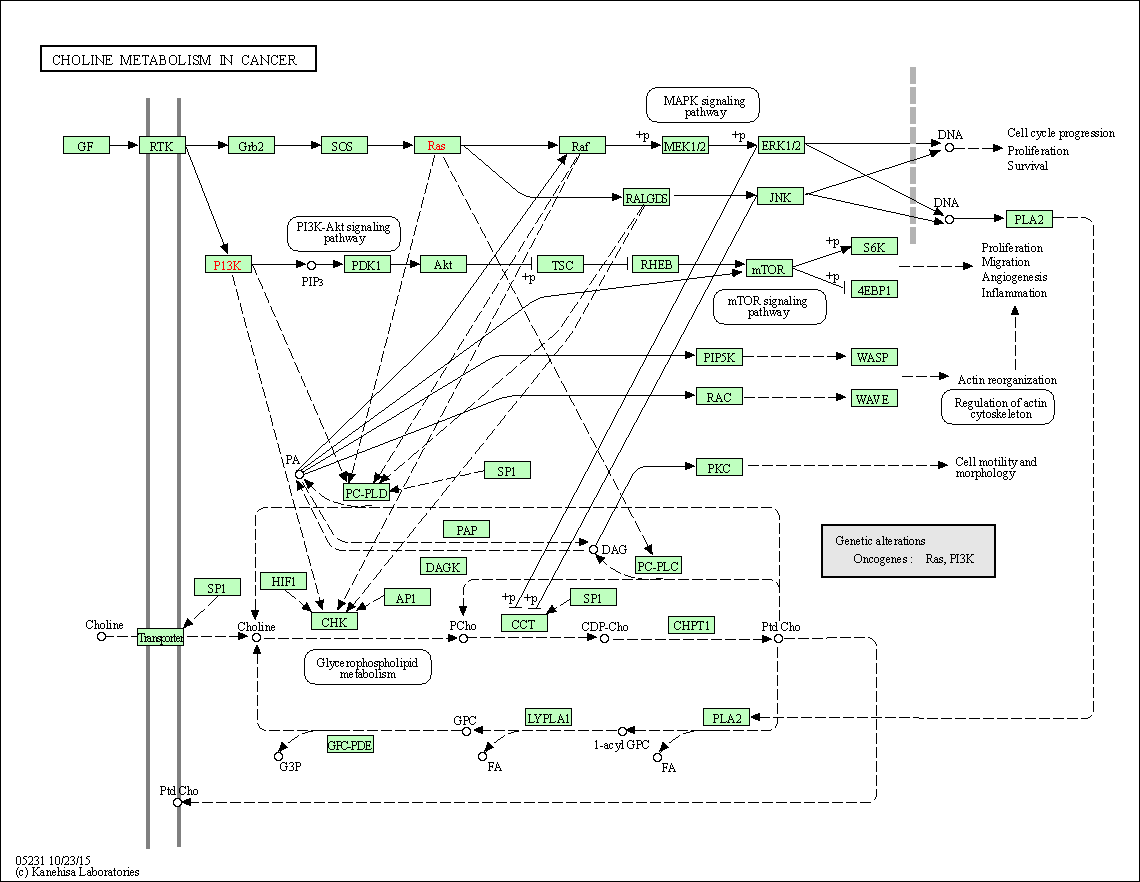
|
|||||||
| PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 |
Pathway Map 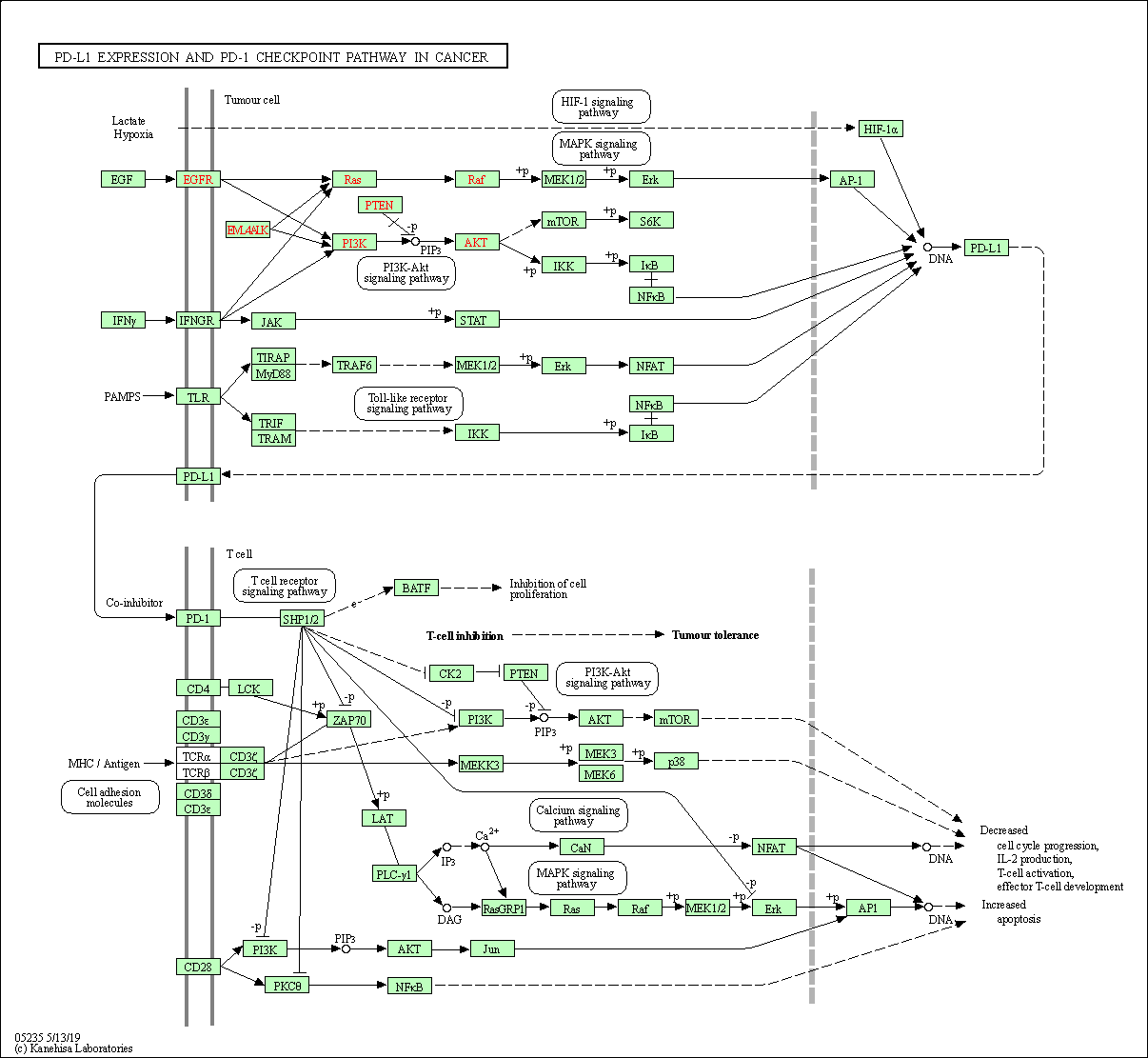
|
|||||||
| Glioma | hsa05214 |
Pathway Map 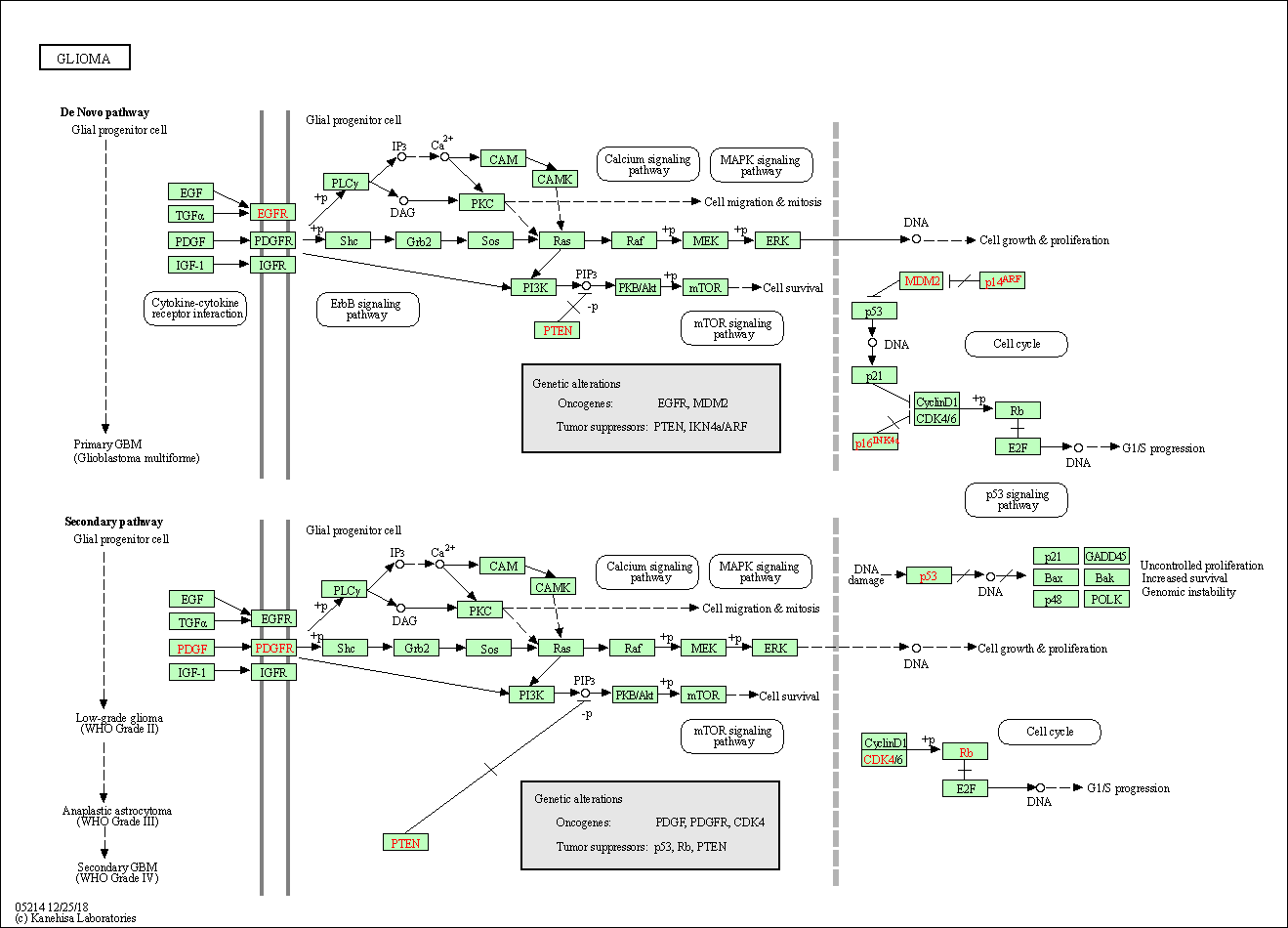
|
|||||||
| Non-small cell lung cancer | hsa05223 |
Pathway Map 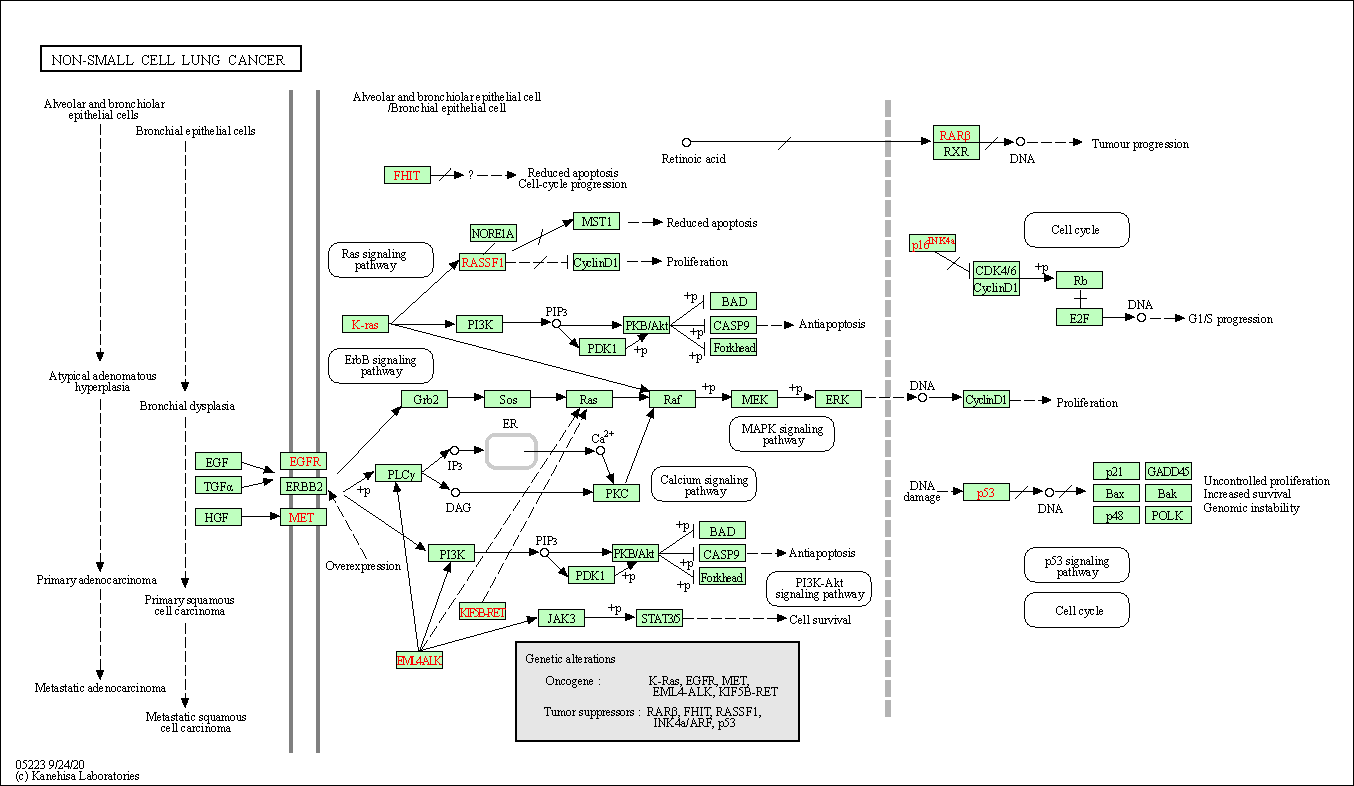
|
|||||||
| Hepatocellular carcinoma | hsa05225 |
Pathway Map 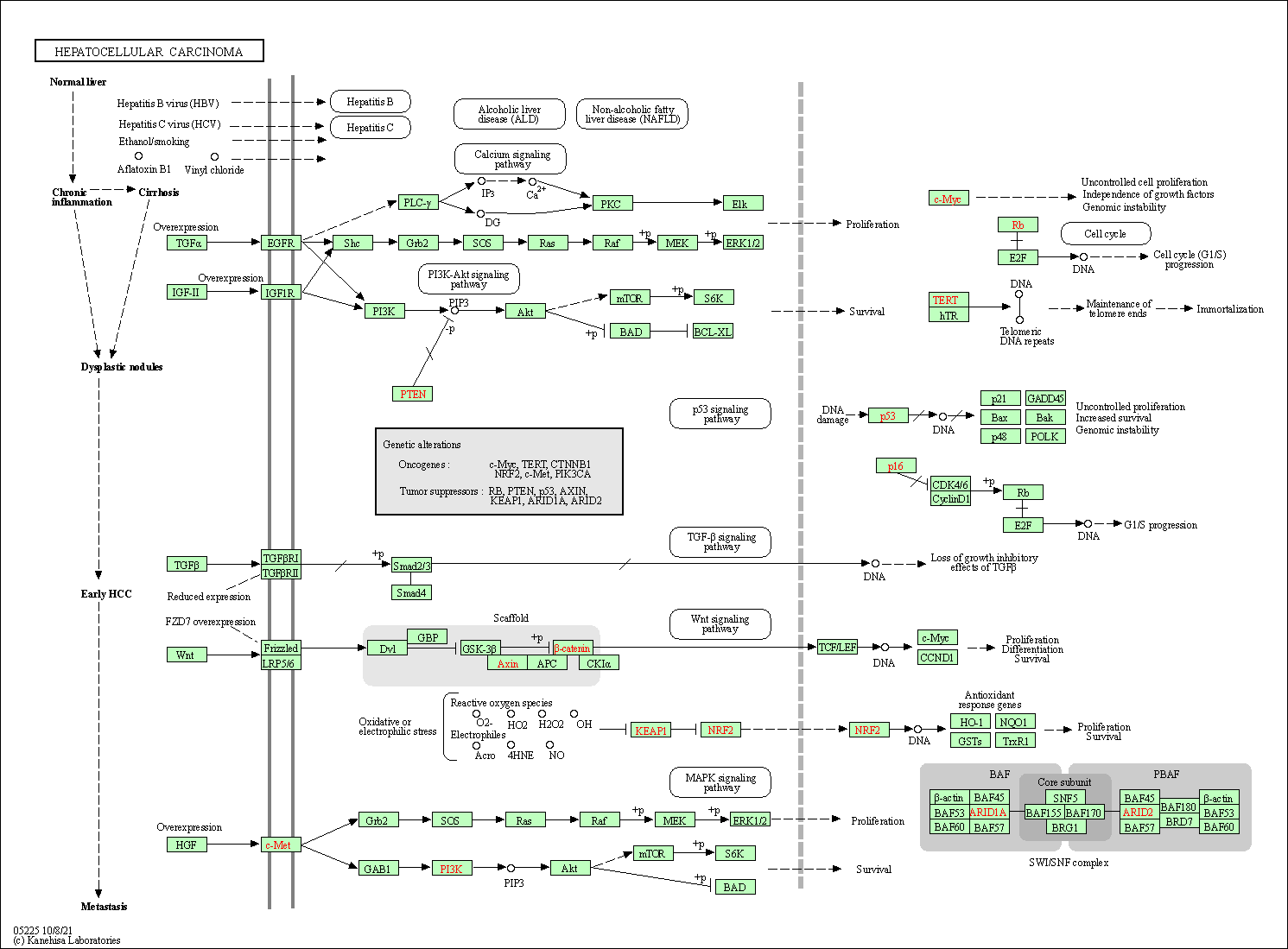
|
|||||||
| Inositol phosphate metabolism | hsa00562 |
Pathway Map 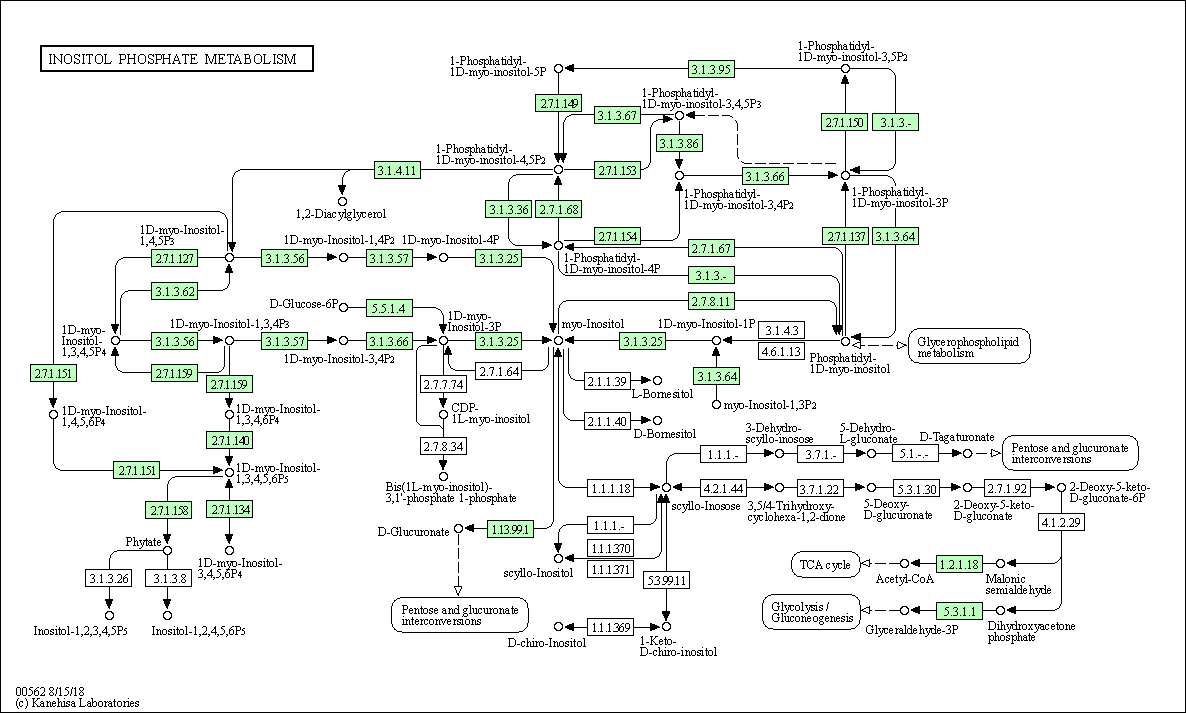
|
|||||||
| Lipid and atherosclerosis | hsa05417 |
Pathway Map 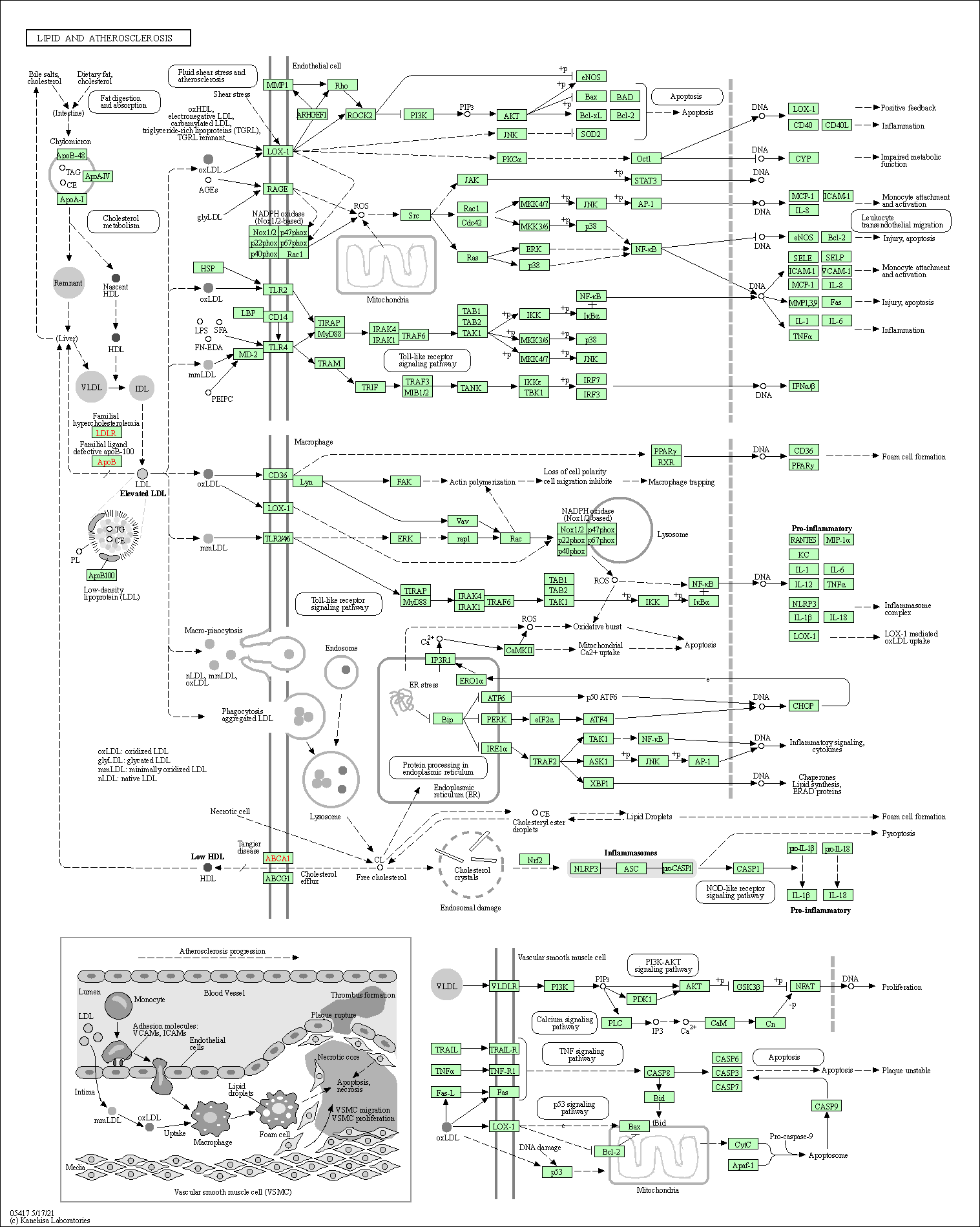
|
|||||||
| Axon guidance | hsa04360 |
Pathway Map 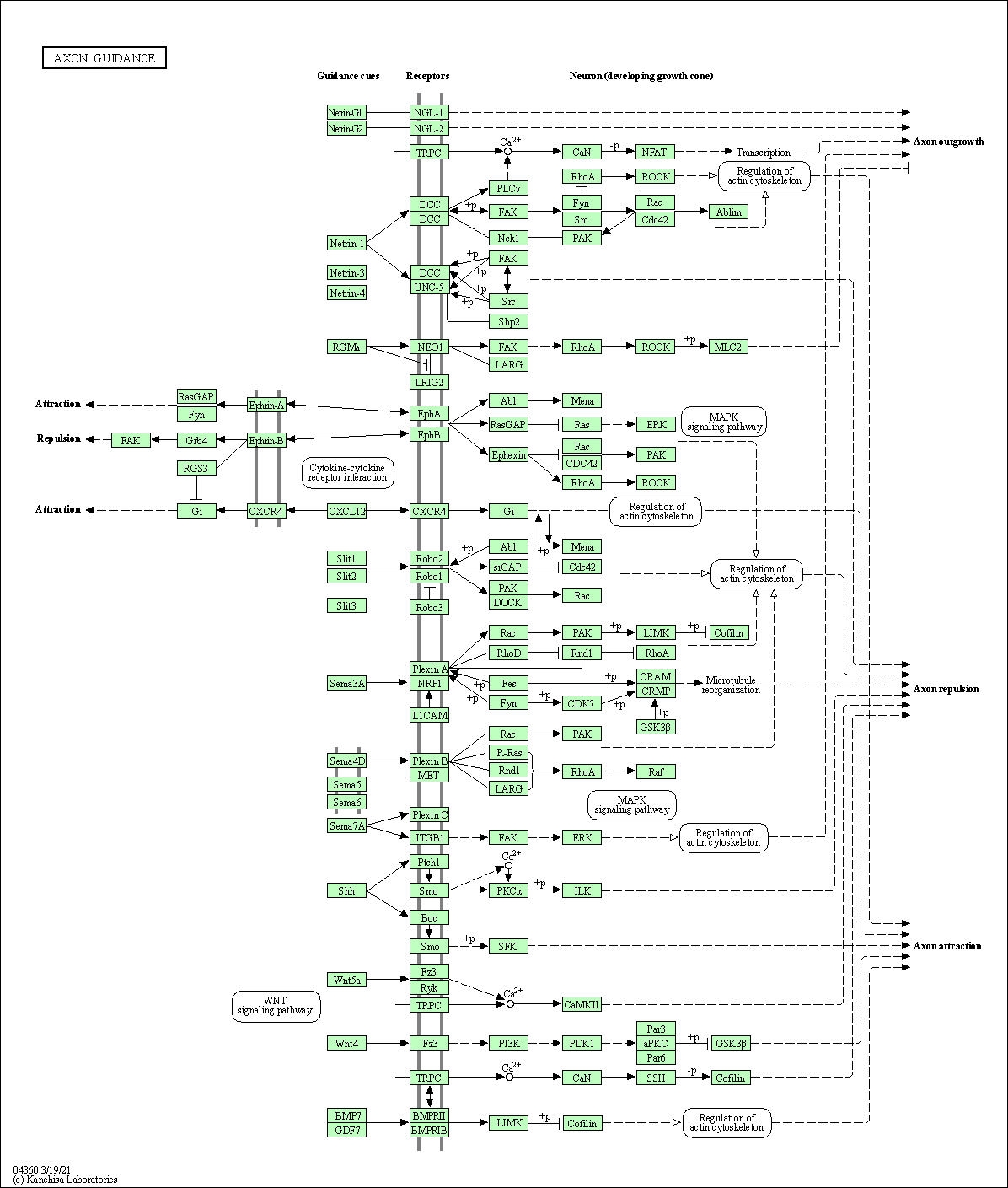
|
|||||||
| AGE-RAGE signaling pathway in diabetic complications | hsa04933 |
Pathway Map 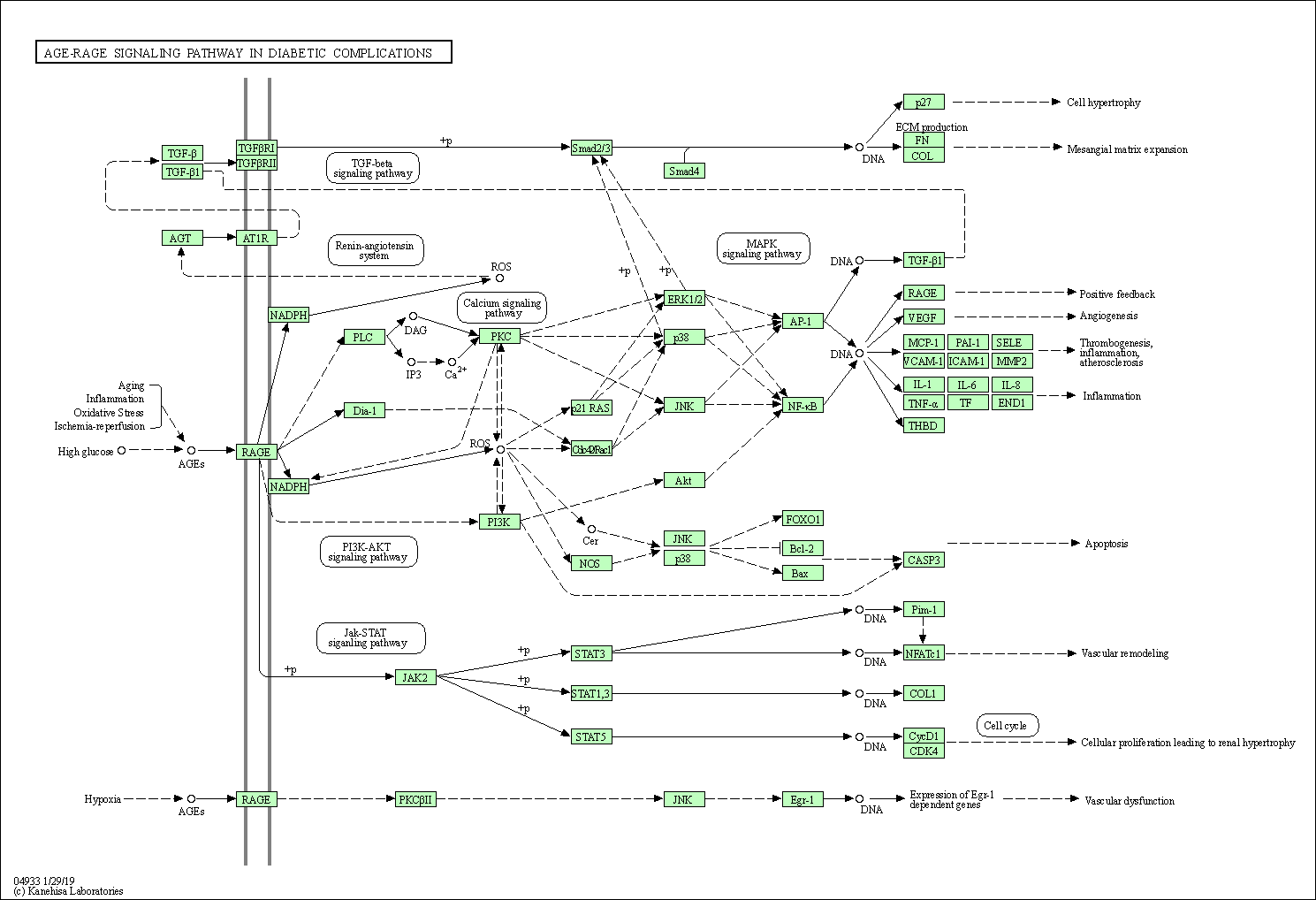
|
|||||||
| Thyroid hormone signaling pathway | hsa04919 |
Pathway Map 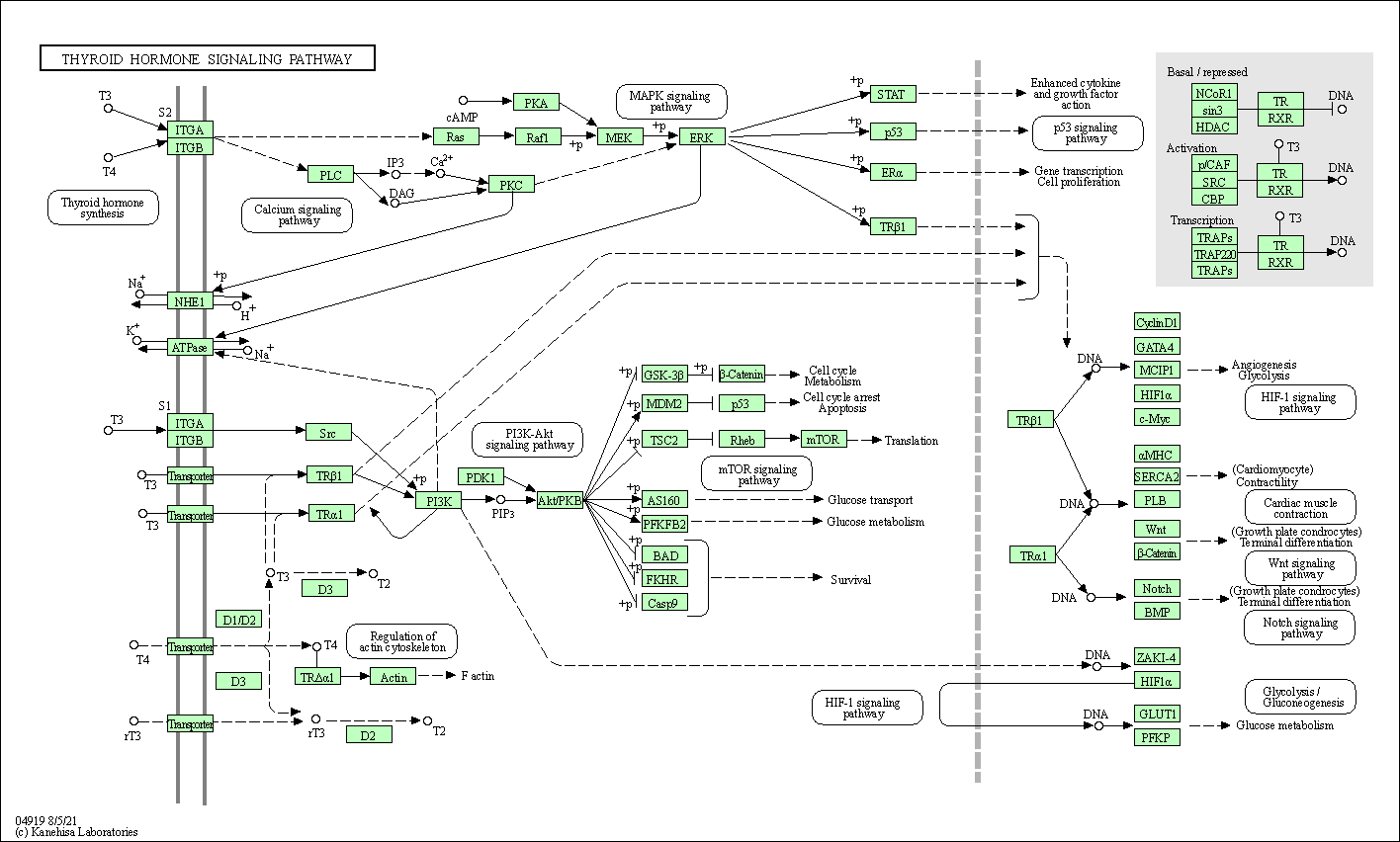
|
|||||||
| Growth hormone synthesis, secretion and action | hsa04935 |
Pathway Map 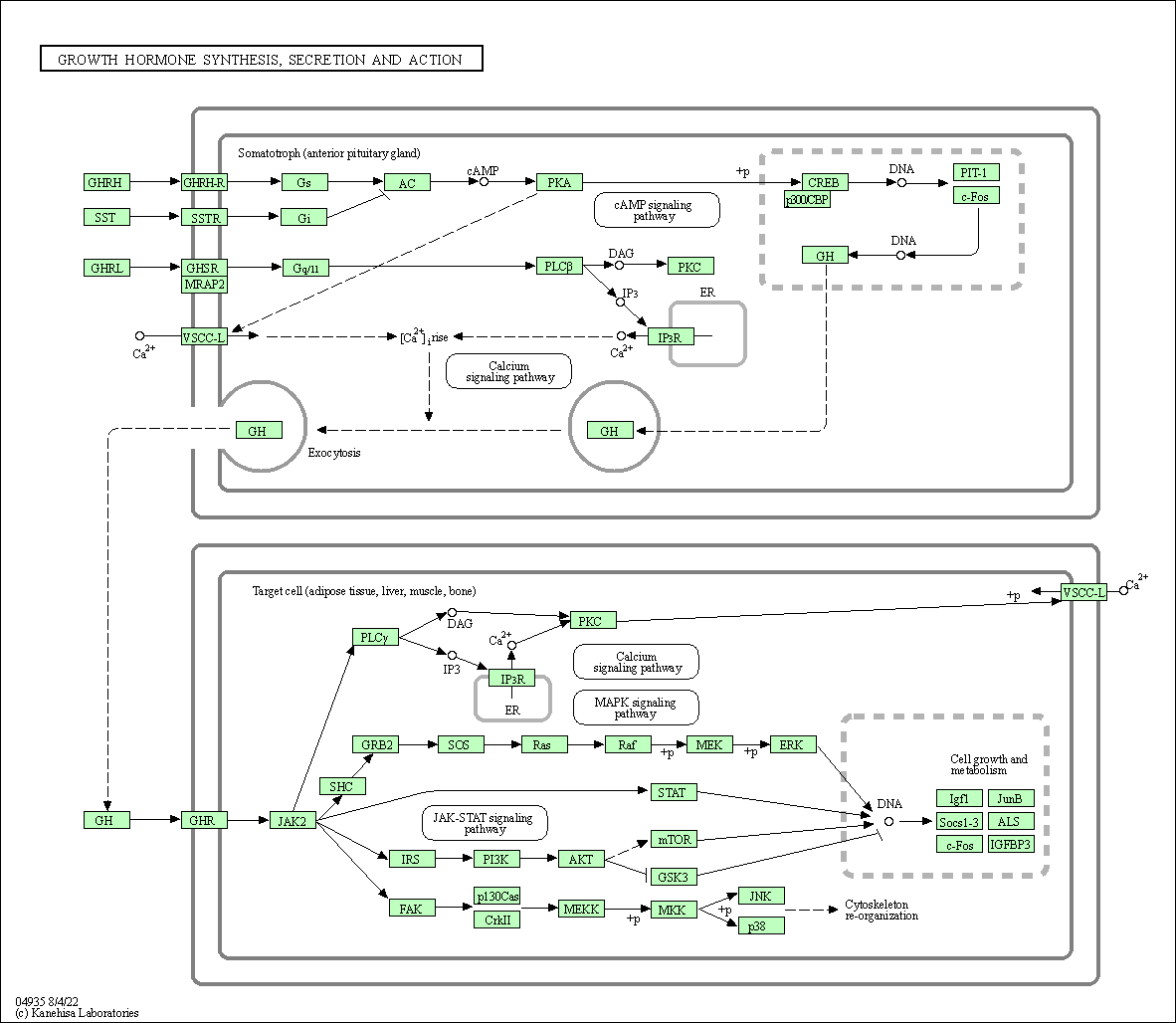
|
|||||||
| Metabolic pathways | hsa01100 |
Pathway Map 
|
|||||||
| Neurotrophin signaling pathway | hsa04722 |
Pathway Map 
|
|||||||
| Parkinson disease | hsa05012 |
Pathway Map 
|
|||||||
| Pathways of neurodegeneration - multiple diseases | hsa05022 |
Pathway Map 
|
|||||||
| Inflammatory mediator regulation of TRP channels | hsa04750 |
Pathway Map 
|
|||||||
| ErbB signaling pathway | hsa04012 |
Pathway Map 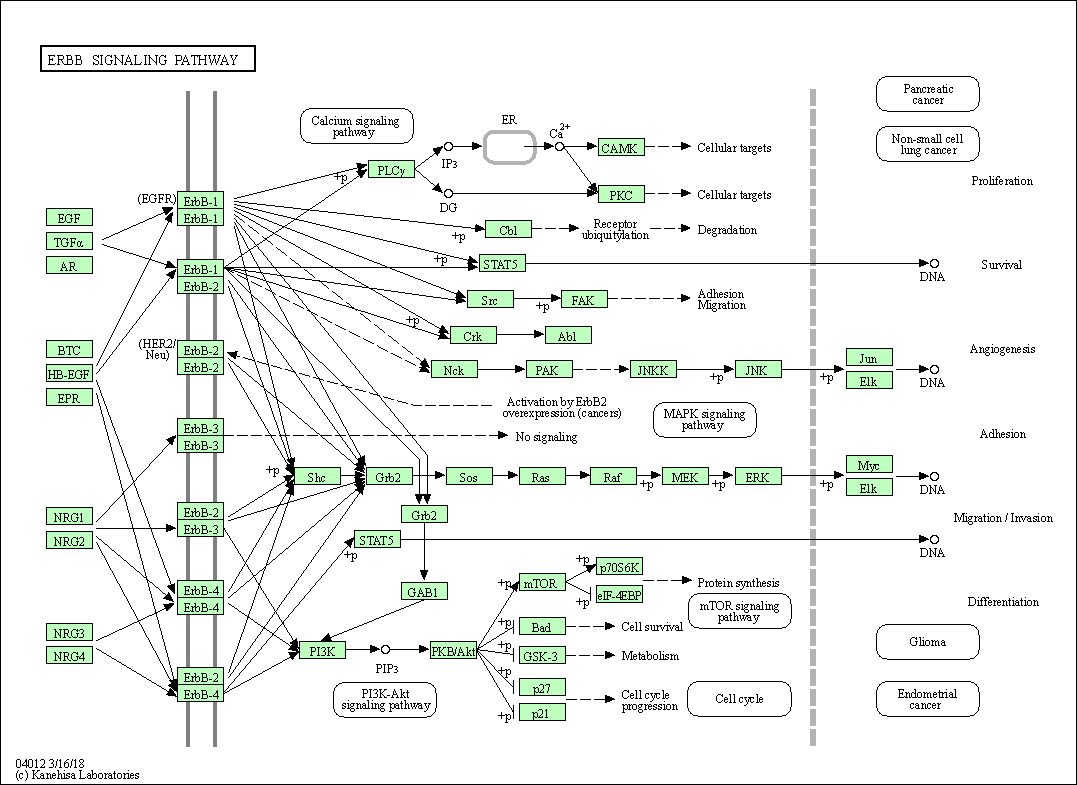
|
|||||||
| Ras signaling pathway | hsa04014 |
Pathway Map 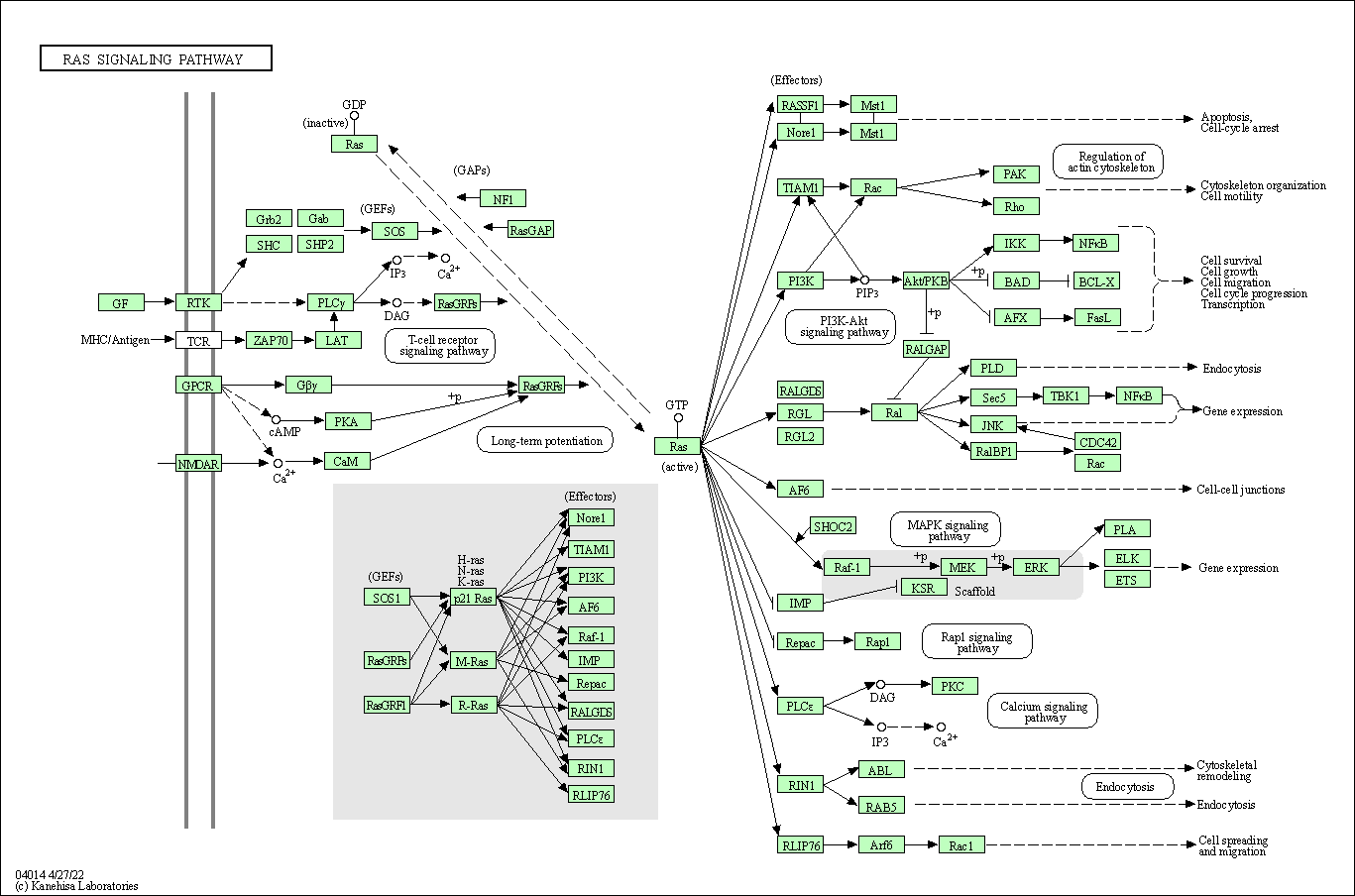
|
|||||||
| Rap1 signaling pathway | hsa04015 |
Pathway Map 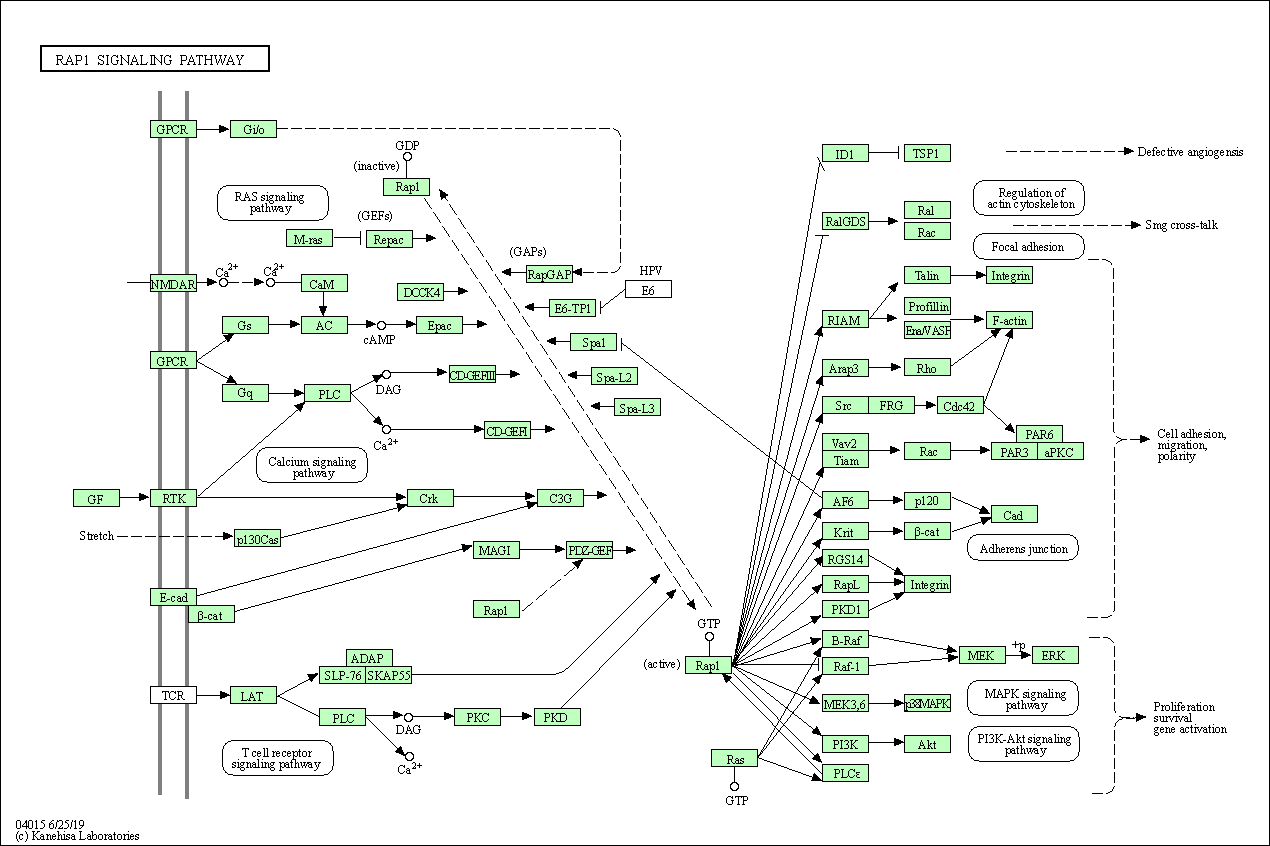
|
|||||||
| Calcium signaling pathway | hsa04020 |
Pathway Map 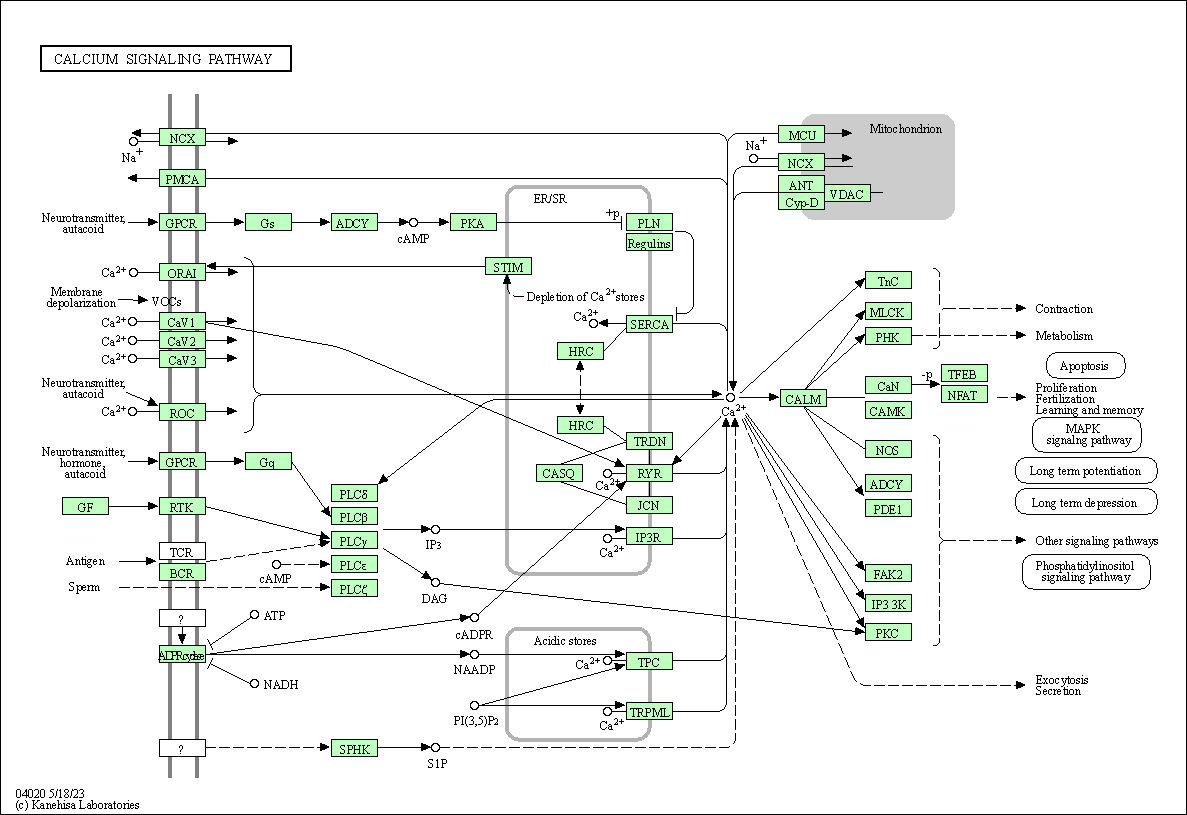
|
|||||||
| NF-kappa B signaling pathway | hsa04064 |
Pathway Map 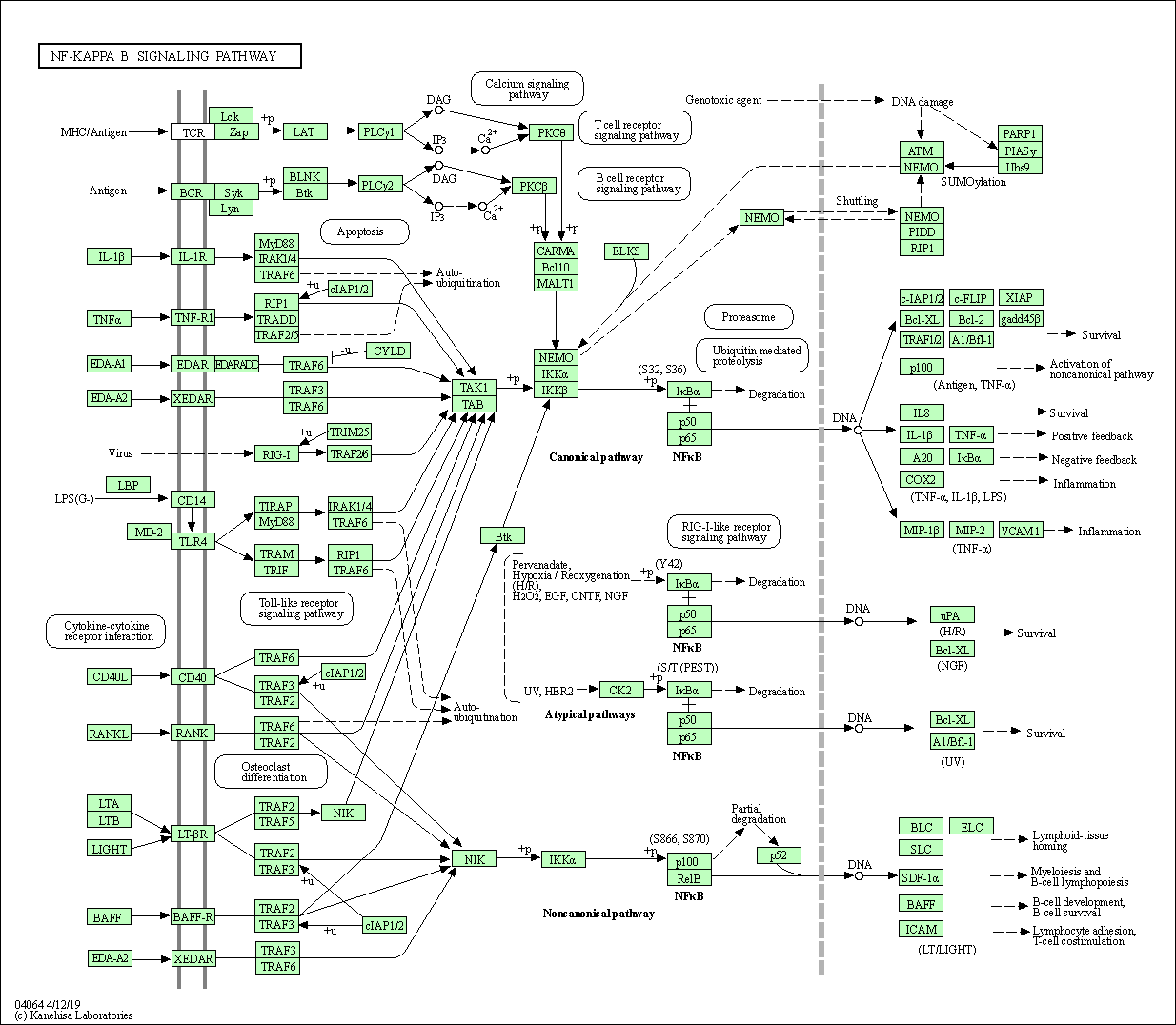
|
|||||||
| HIF-1 signaling pathway | hsa04066 |
Pathway Map 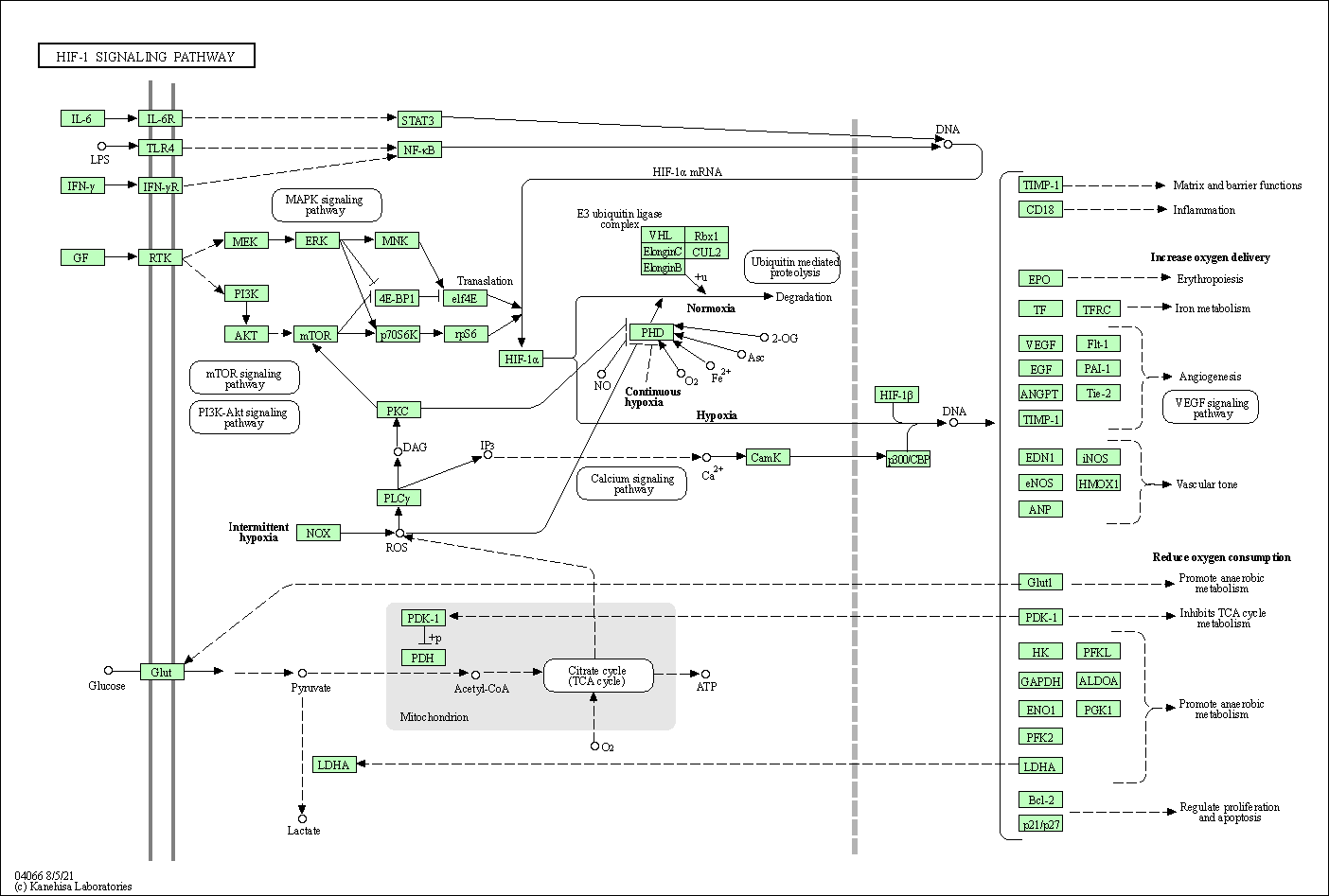
|
|||||||
| Phosphatidylinositol signaling system | hsa04070 |
Pathway Map 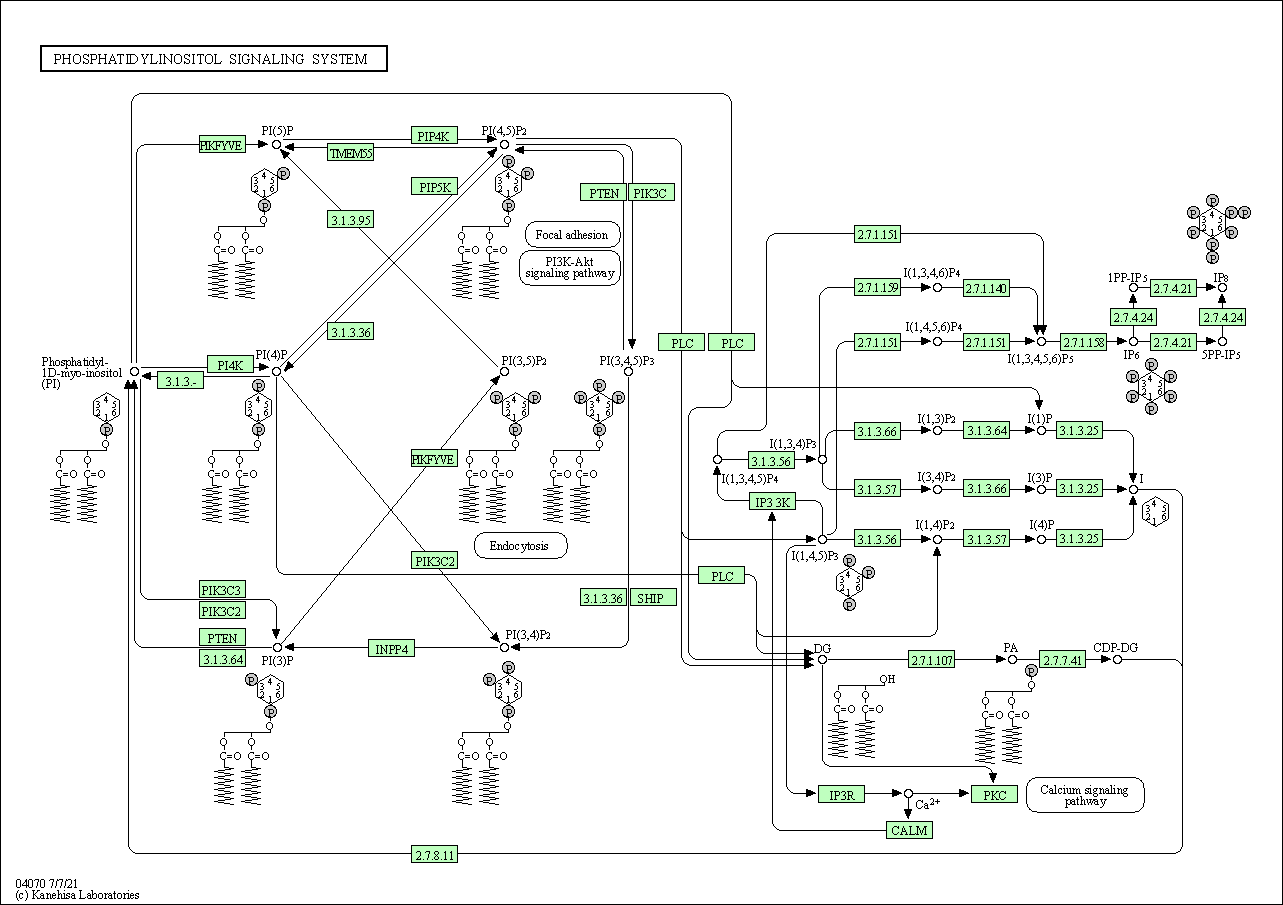
|
|||||||
| Phospholipase D signaling pathway | hsa04072 |
Pathway Map 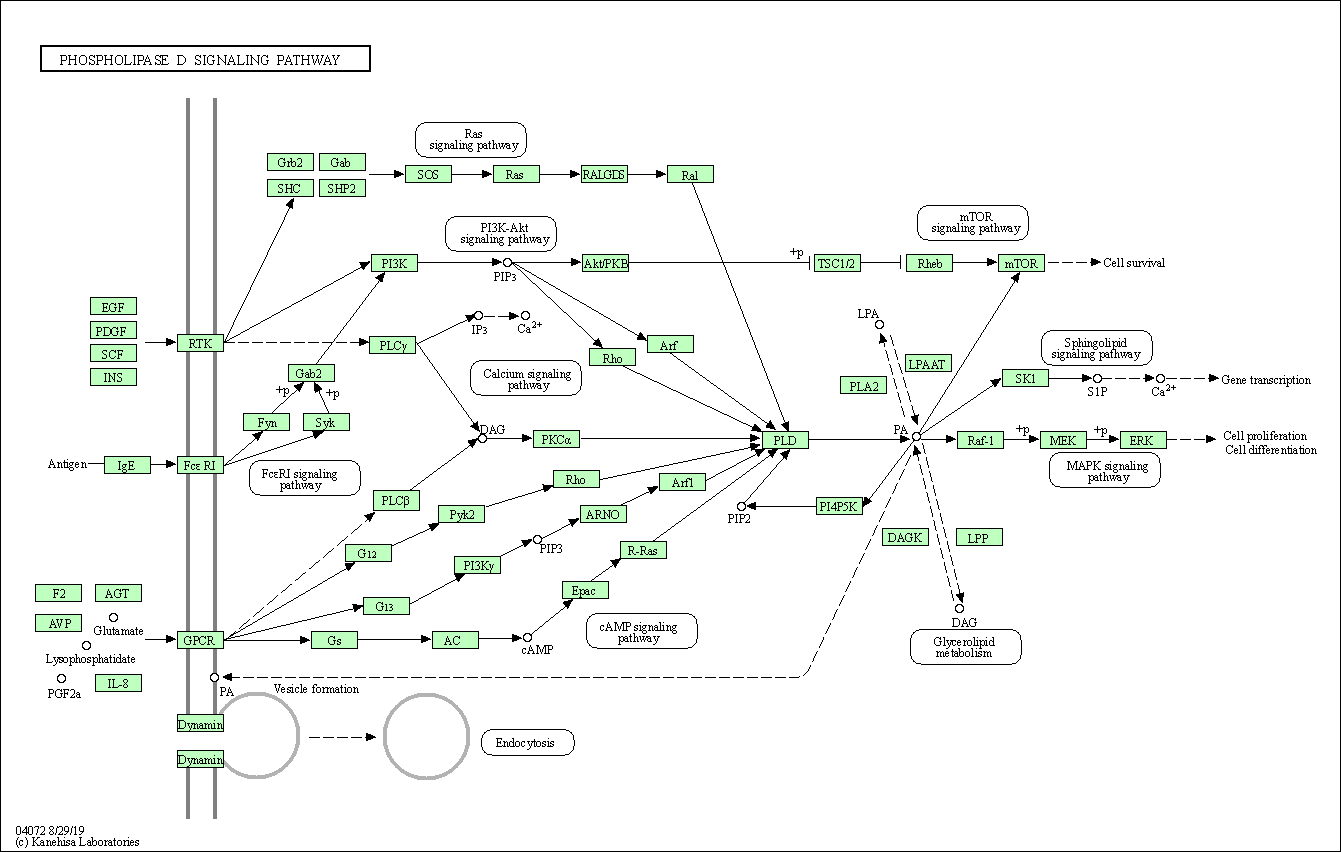
|
|||||||
| VEGF signaling pathway | hsa04370 |
Pathway Map 
|
|||||||
| 3D Structure |
|
||||||||
Full List of Virus RNA Interacting with This Protien
| Virus Name | Binding Region | RNA Info | Detection Method | Infection Cell | Cell ID | Cell Originated Tissue | Infection Time | Interaction Score 1 | Interaction Score 2 |
|---|---|---|---|---|---|---|---|---|---|
| Chikungunya virus (strain 181/25) | 5'UTR - 3'UTR | RNA Info | VIRal Cross-Linking And Solid-phase Purification (VIR-CLASP) | BHK-21 Cells (Small hamster kidney fibroblast) | . | Kidney | 0.2 h | P = 0.0590680899981436 | . |
| Chikungunya virus (strain 181/25) | 5'UTR - 3'UTR | RNA Info | VIRal Cross-Linking And Solid-phase Purification (VIR-CLASP) | BHK-21 Cells (Small hamster kidney fibroblast) | . | Kidney | 1 h | P = 0.0465317995661615 | . |
| Chikungunya virus (strain 181/25) | 5'UTR - 3'UTR | RNA Info | VIRal Cross-Linking And Solid-phase Purification (VIR-CLASP) | BHK-21 Cells (Small hamster kidney fibroblast) | . | Kidney | 3 h | P = 0.00154551109133323 | . |
Potential Drug(s) that Targets This Protein
Protein Sequence Information
|
MAGAASPCANGCGPGAPSDAEVLHLCRSLEVGTVMTLFYSKKSQRPERKTFQVKLETRQITWSRGADKIEGAIDIREIKEIRPGKTSRDFDRYQEDPAFRPDQSHCFVILYGMEFRLKTLSLQATSEDEVNMWIKGLTWLMEDTLQAPTPLQIERWLRKQFYSVDRNREDRISAKDLKNMLSQVNYRVPNMRFLRERLTDLEQRSGDITYGQFAQLYRSLMYSAQKTMDLPFLEASTLRAGERPELCRVSLPEFQQFLLDYQGELWAVDRLQVQEFMLSFLRDPLREIEEPYFFLDEFVTFLFSKENSVWNSQLDAVCPDTMNNPLSHYWISSSHNTYLTGDQFSSESSLEAYARCLRMGCRCIELDCWDGPDGMPVIYHGHTLTTKIKFSDVLHTIKEHAFVASEYPVILSIEDHCSIAQQRNMAQYFKKVLGDTLLTKPVEISADGLPSPNQLKRKILIKHKKLAEGSAYEEVPTSMMYSENDISNSIKNGILYLEDPVNHEWYPHYFVLTSSKIYYSEETSSDQGNEDEEEPKEVSSSTELHSNEKWFHGKLGAGRDGRHIAERLLTEYCIETGAPDGSFLVRESETFVGDYTLSFWRNGKVQHCRIHSRQDAGTPKFFLTDNLVFDSLYDLITHYQQVPLRCNEFEMRLSEPVPQTNAHESKEWYHASLTRAQAEHMLMRVPRDGAFLVRKRNEPNSYAISFRAEGKIKHCRVQQEGQTVMLGNSEFDSLVDLISYYEKHPLYRKMKLRYPINEEALEKIGTAEPDYGALYEGRNPGFYVEANPMPTFKCAVKALFDYKAQREDELTFIKSAIIQNVEKQEGGWWRGDYGGKKQLWFPSNYVEEMVNPVALEPEREHLDENSPLGDLLRGVLDVPACQIAIRPEGKNNRLFVFSISMASVAHWSLDVAADSQEELQDWVKKIREVAQTADARLTEGKIMERRKKIALELSELVVYCRPVPFDEEKIGTERACYRDMSSFPETKAEKYVNKAKGKKFLQYNRLQLSRIYPKGQRLDSSNYDPLPMWICGSQLVALNFQTPDKPMQMNQALFMTGRHCGYVLQPSTMRDEAFDPFDKSSLRGLEPCAISIEVLGARHLPKNGRGIVCPFVEIEVAGAEYDSTKQKTEFVVDNGLNPVWPAKPFHFQISNPEFAFLRFVVYEEDMFSDQNFLAQATFPVKGLKTGYRAVPLKNNYSEDLELASLLIKIDIFPAKENGDLSPFSGTSLRERGSDASGQLFHGRAREGSFESRYQQPFEDFRISQEHLADHFDSRERRAPRRTRVNGDNRL
Click to Show/Hide
|
