Host Protein General Information
| Protein Name |
Actin
|
Gene Name |
ACTB
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Host Species |
Homo sapiens
|
Uniprot Entry Name |
ACTB_HUMAN
|
||||||
| Protein Families |
Actin family
|
||||||||
| EC Number |
3.6.4.-
|
||||||||
| Subcellular Location |
Cytoplasm; cytoskeleton
|
||||||||
| External Link | |||||||||
| NCBI Gene ID | |||||||||
| Uniprot ID | |||||||||
| Ensembl ID | |||||||||
| HGNC ID | |||||||||
| Related KEGG Pathway | |||||||||
| Platelet activation | hsa04611 |
Pathway Map 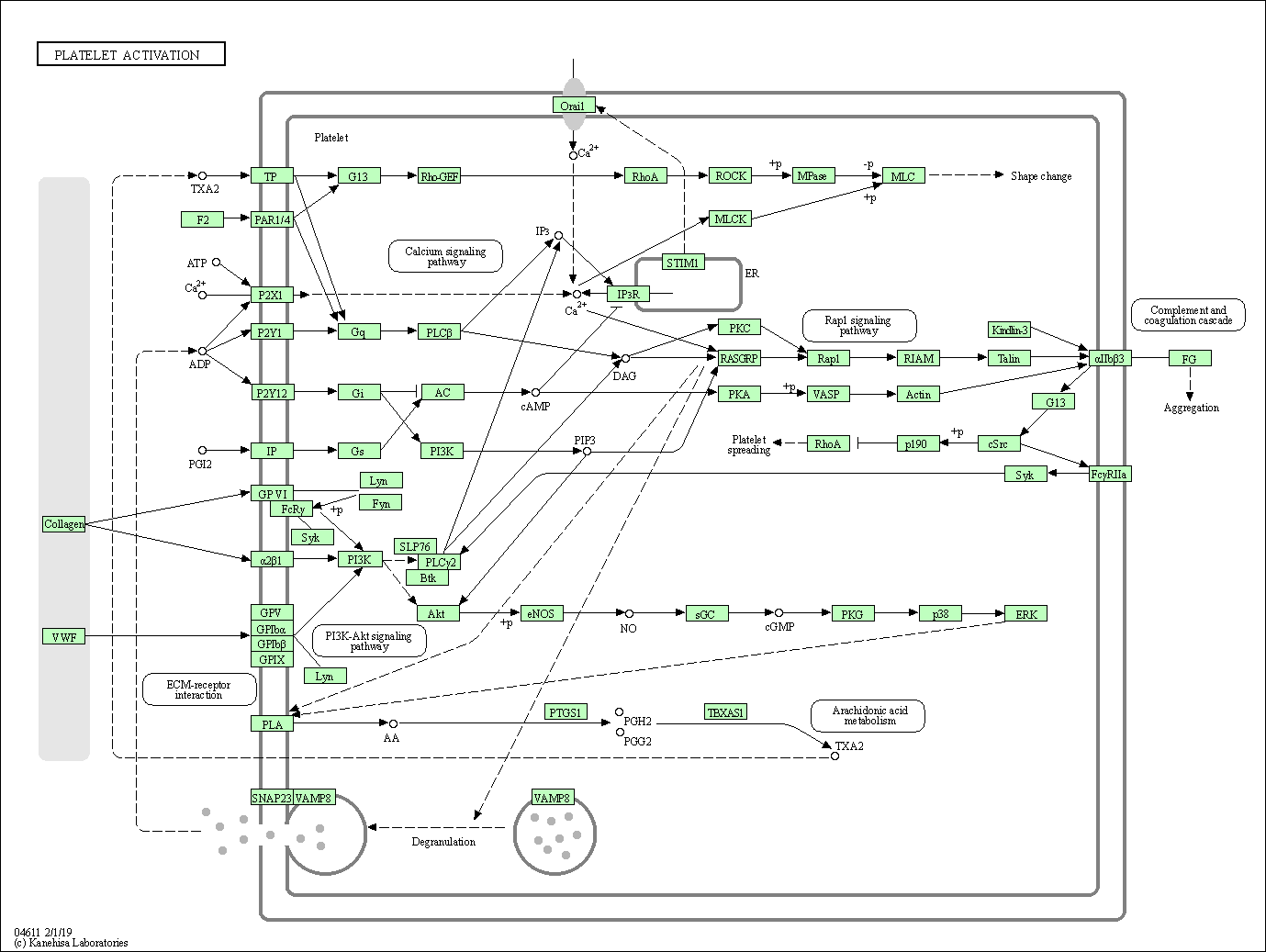
|
|||||||
| Neutrophil extracellular trap formation | hsa04613 |
Pathway Map 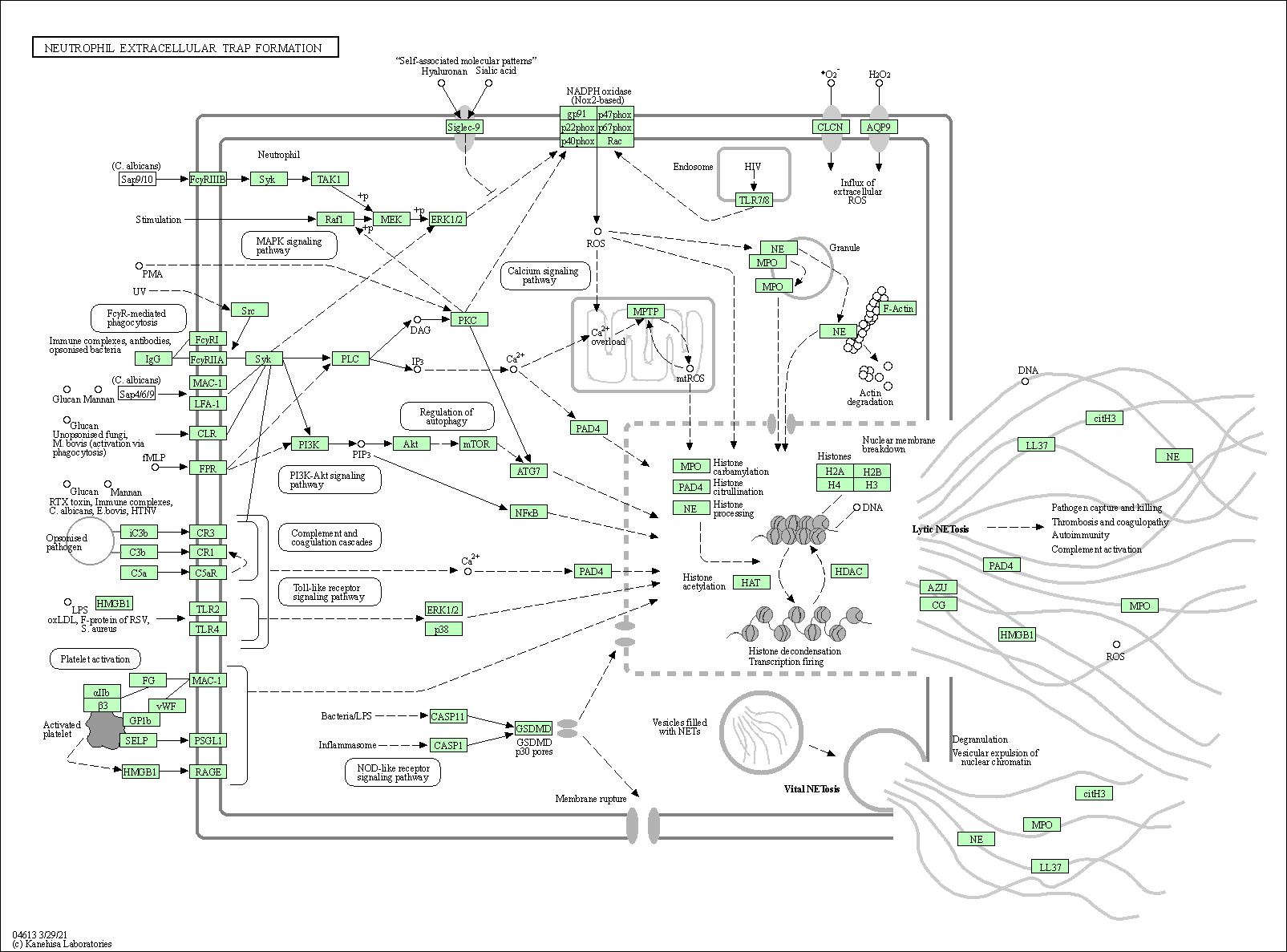
|
|||||||
| Leukocyte transendothelial migration | hsa04670 |
Pathway Map 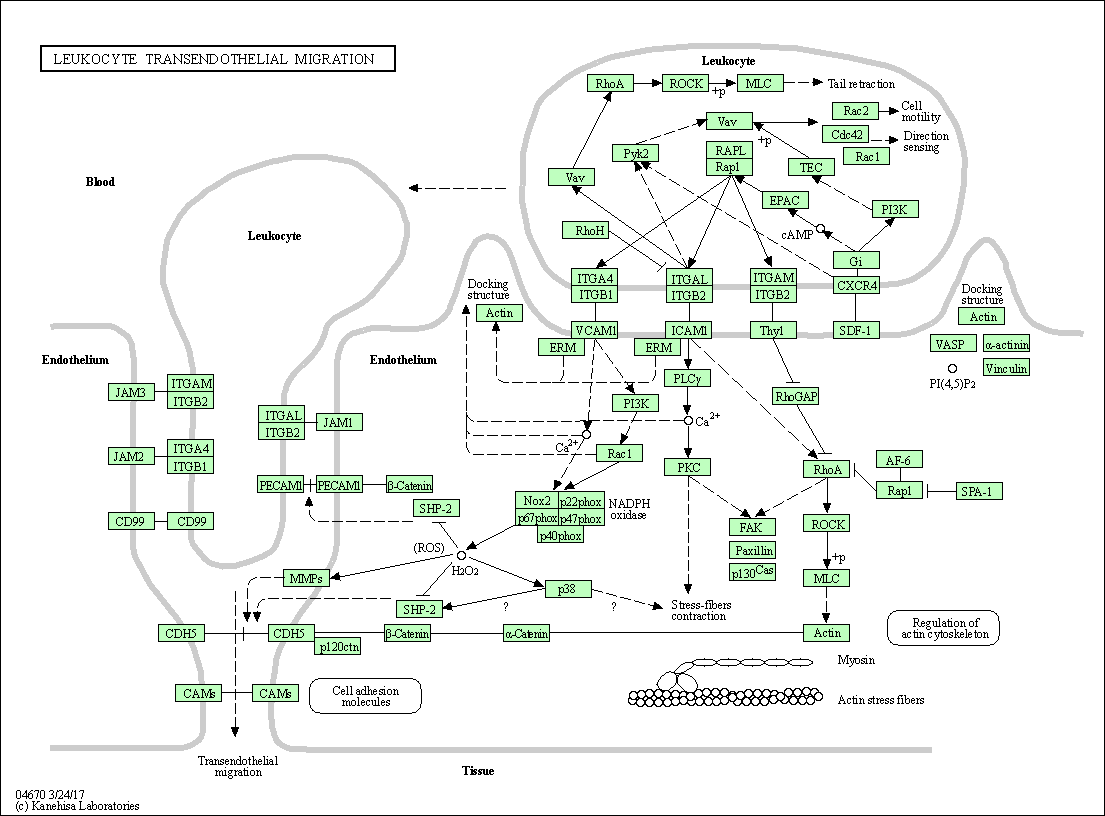
|
|||||||
| Bacterial invasion of epithelial cells | hsa05100 |
Pathway Map 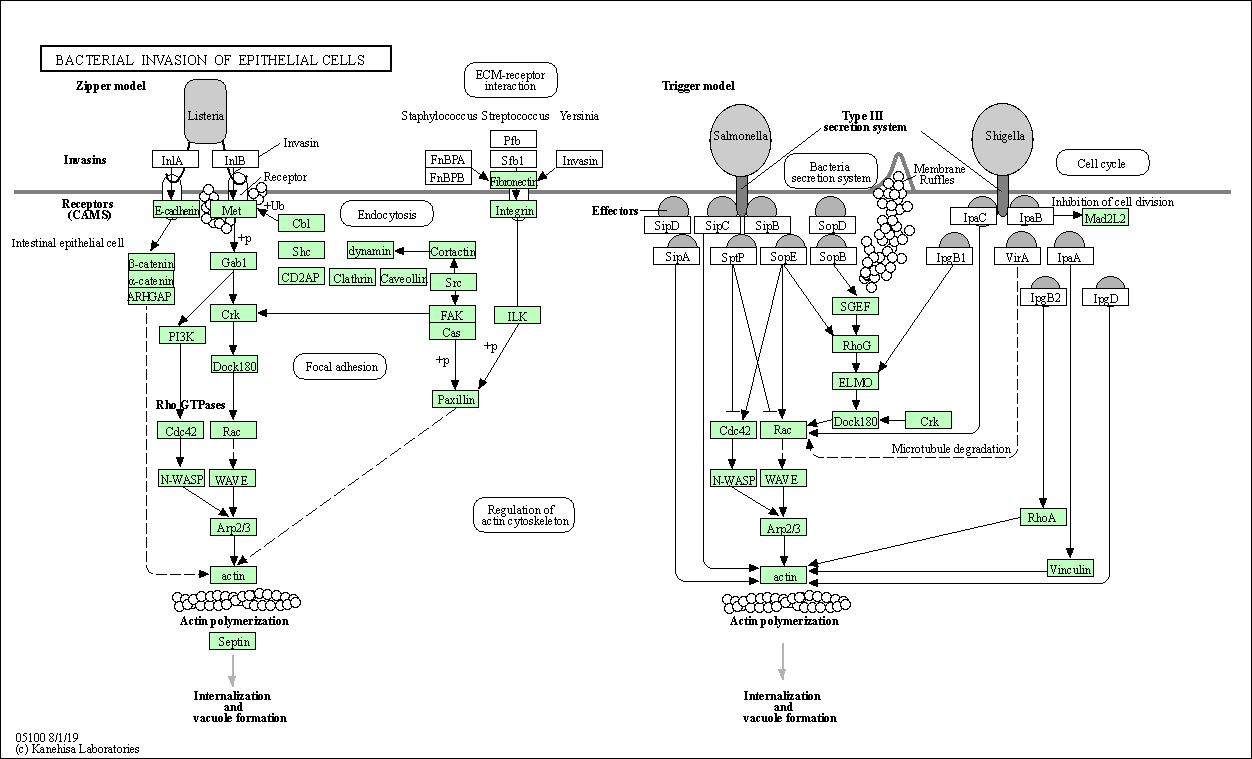
|
|||||||
| Vibrio cholerae infection | hsa05110 |
Pathway Map 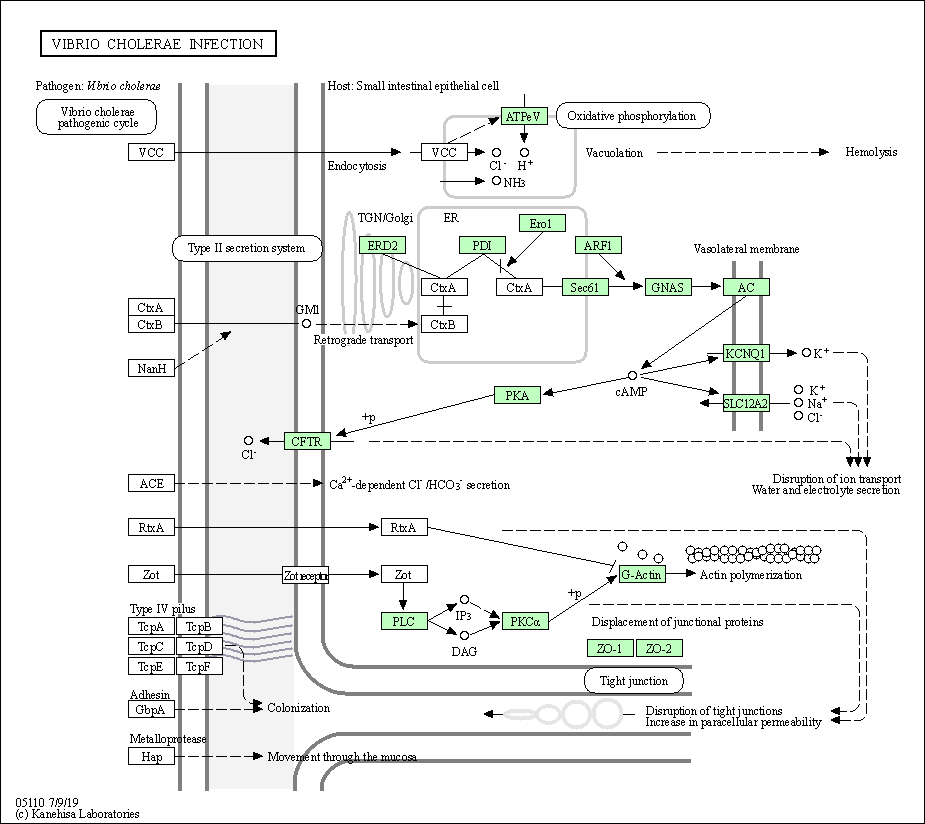
|
|||||||
| Pathogenic Escherichia coli infection | hsa05130 |
Pathway Map 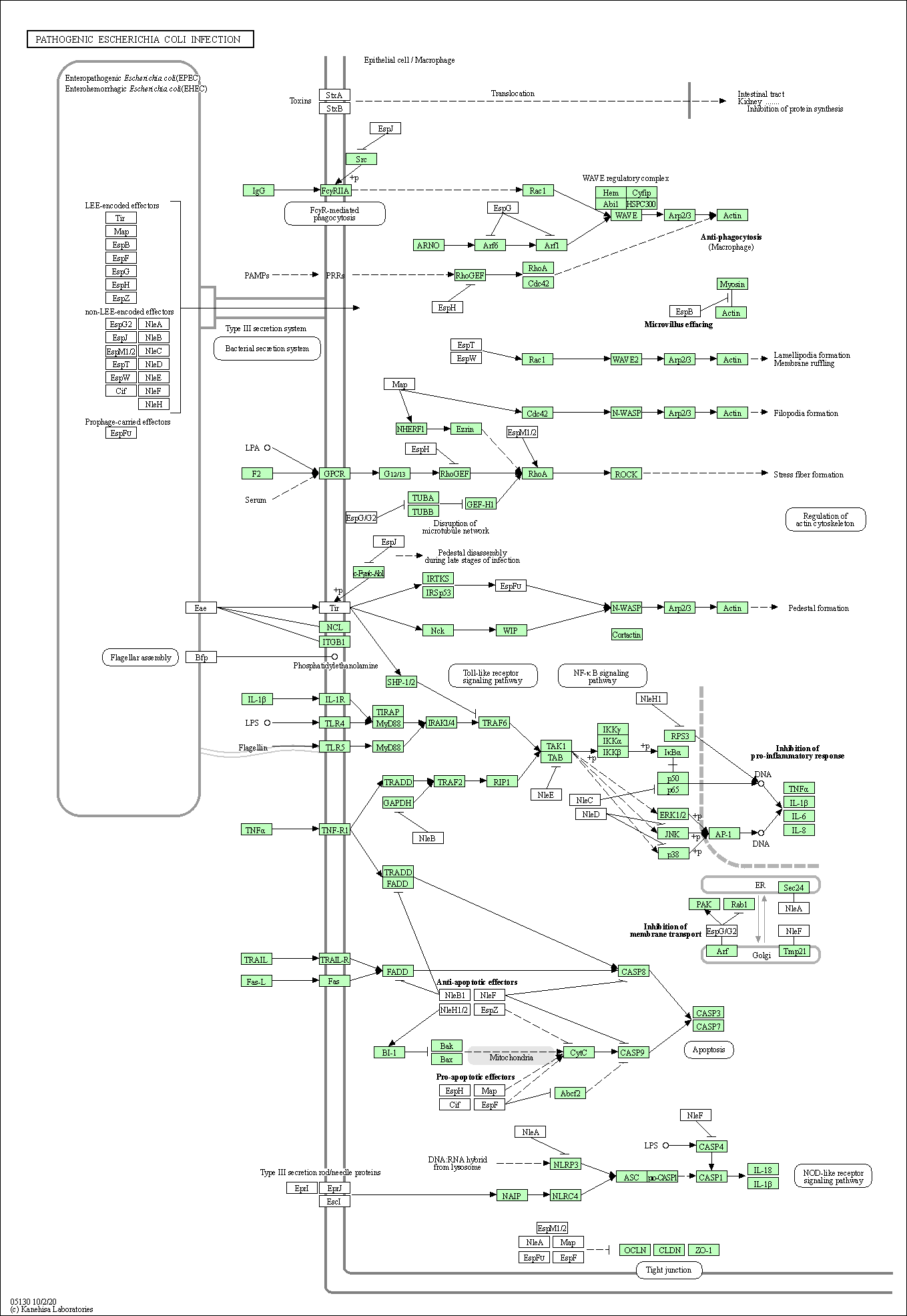
|
|||||||
| Shigellosis | hsa05131 |
Pathway Map 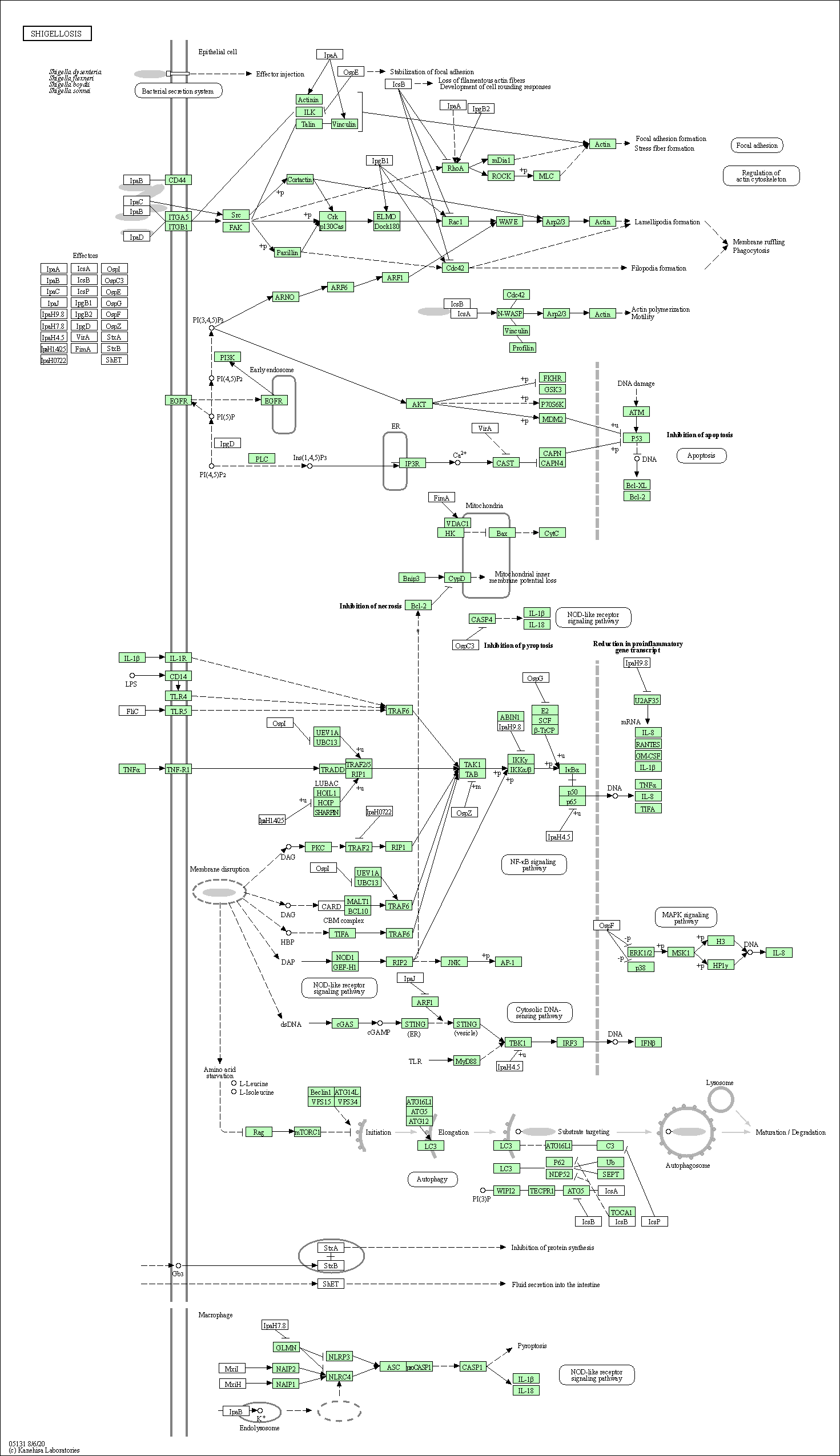
|
|||||||
| Salmonella infection | hsa05132 |
Pathway Map 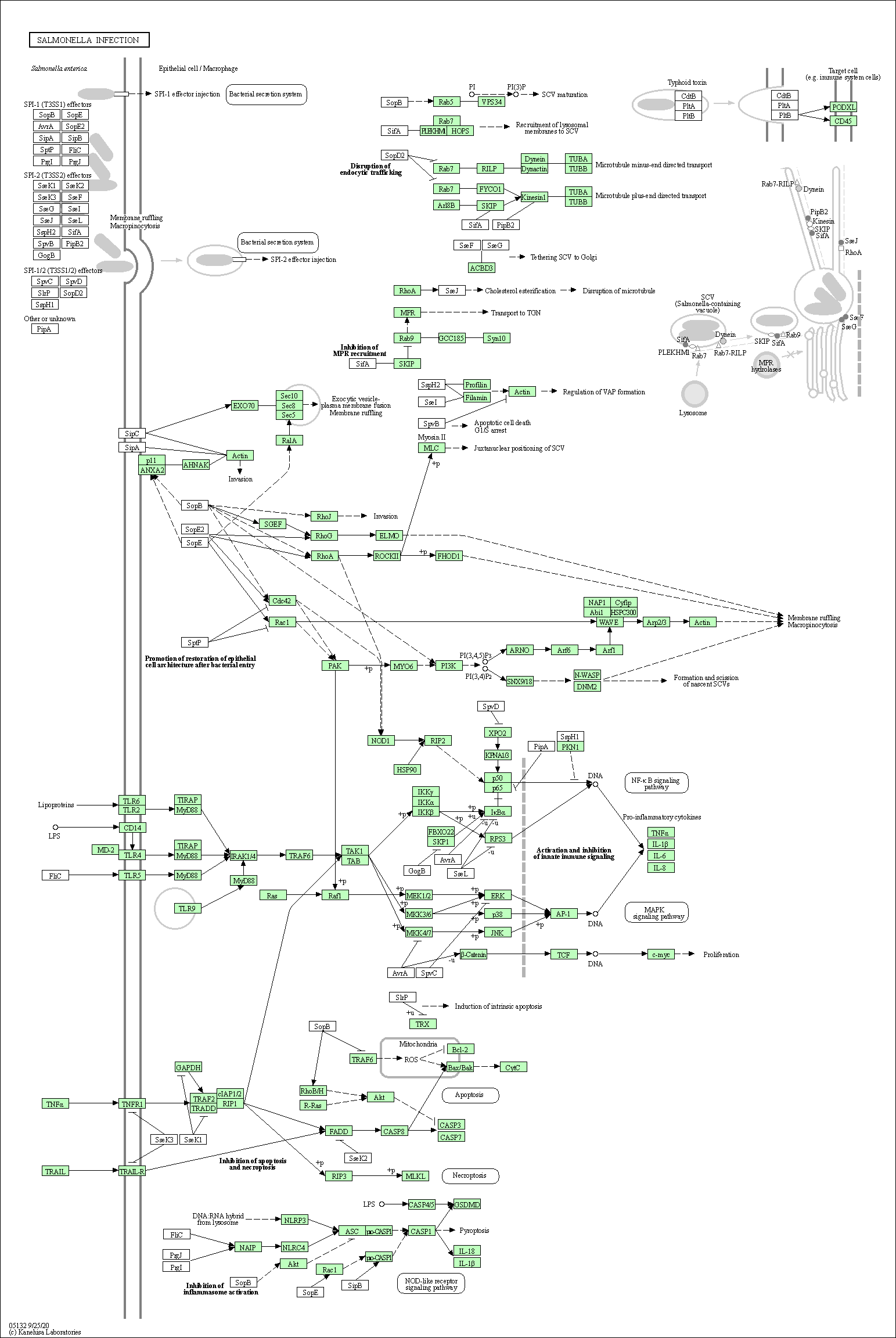
|
|||||||
| Yersinia infection | hsa05135 |
Pathway Map 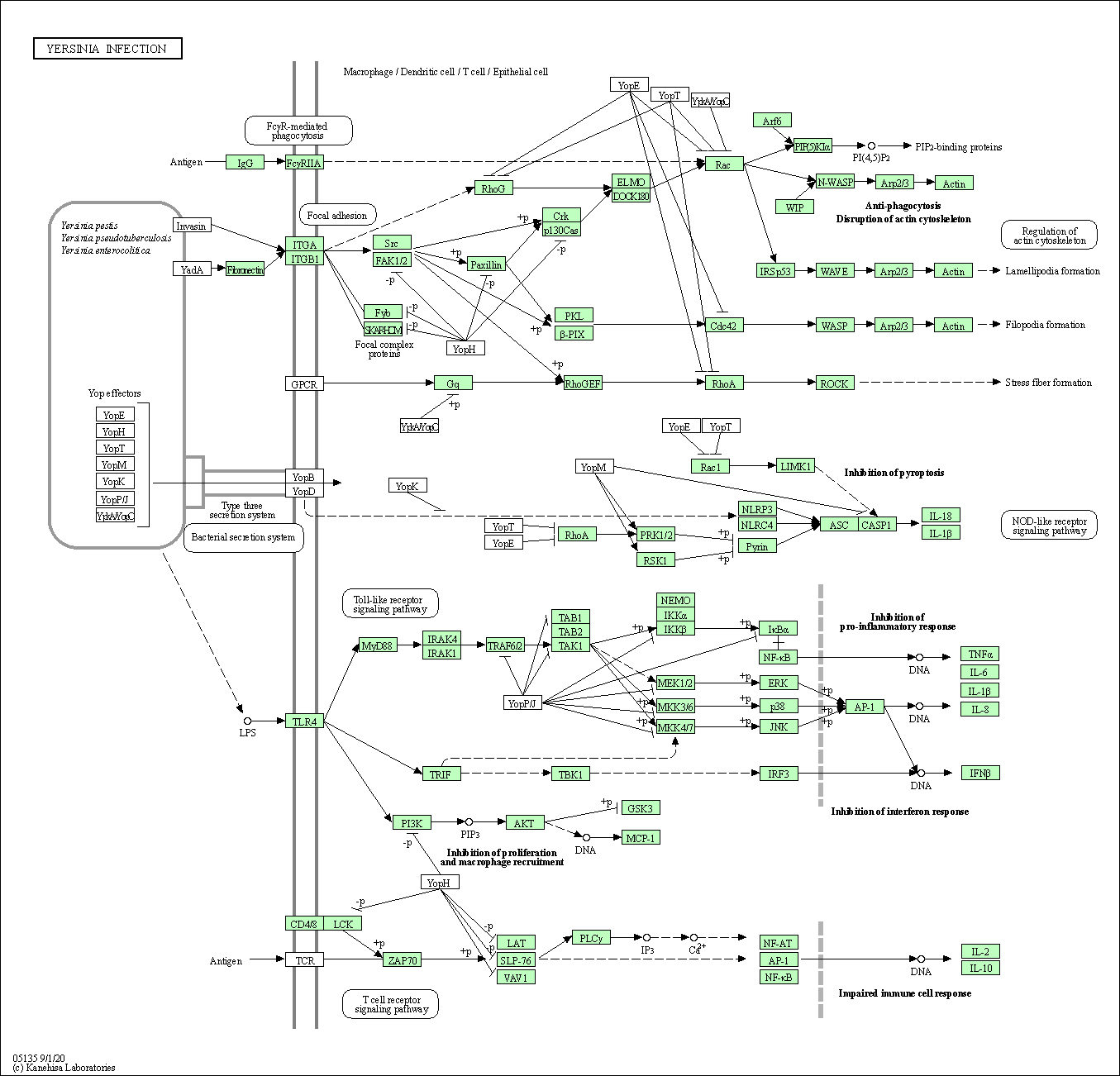
|
|||||||
| Influenza A | hsa05164 |
Pathway Map 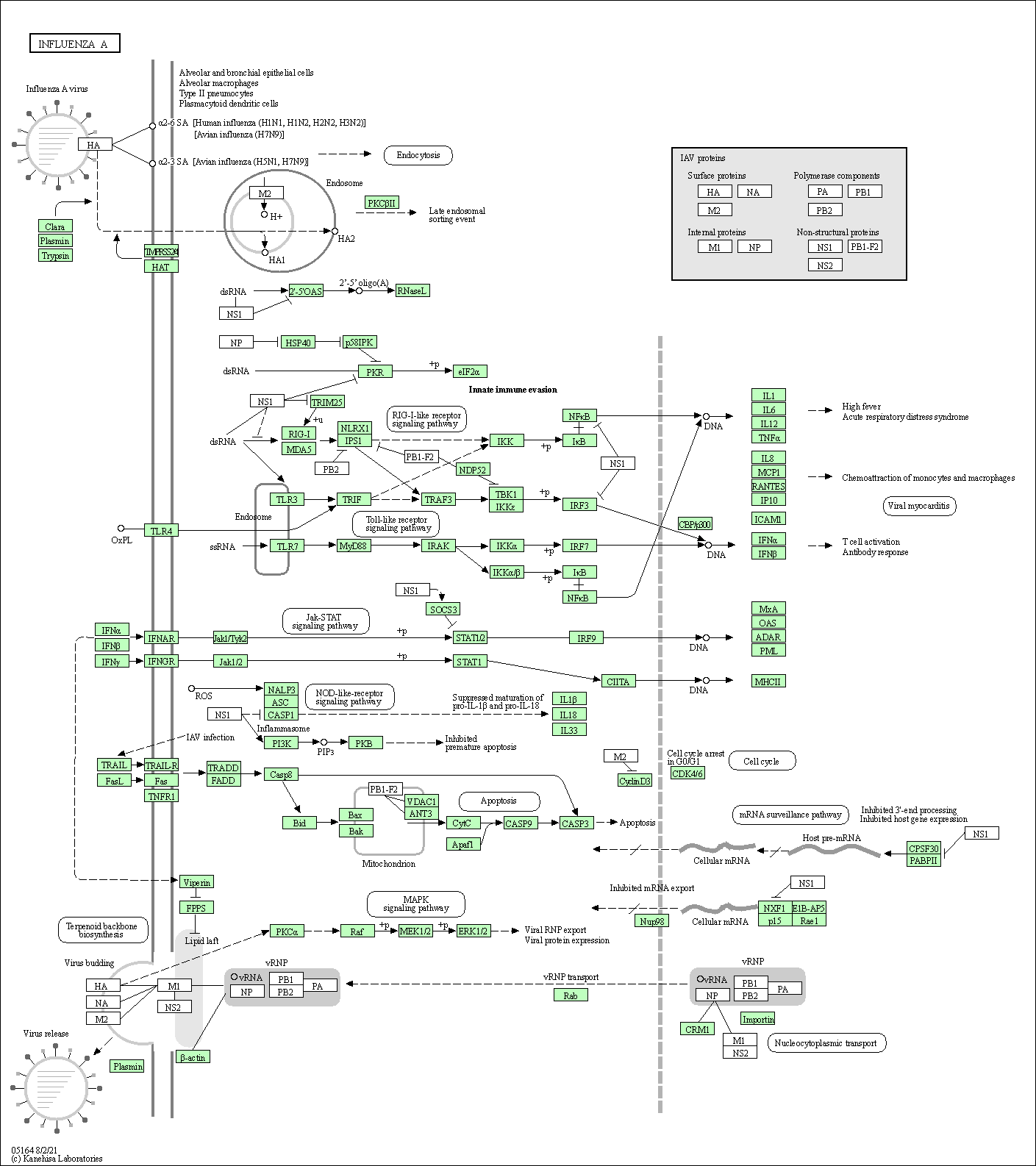
|
|||||||
| Proteoglycans in cancer | hsa05205 |
Pathway Map 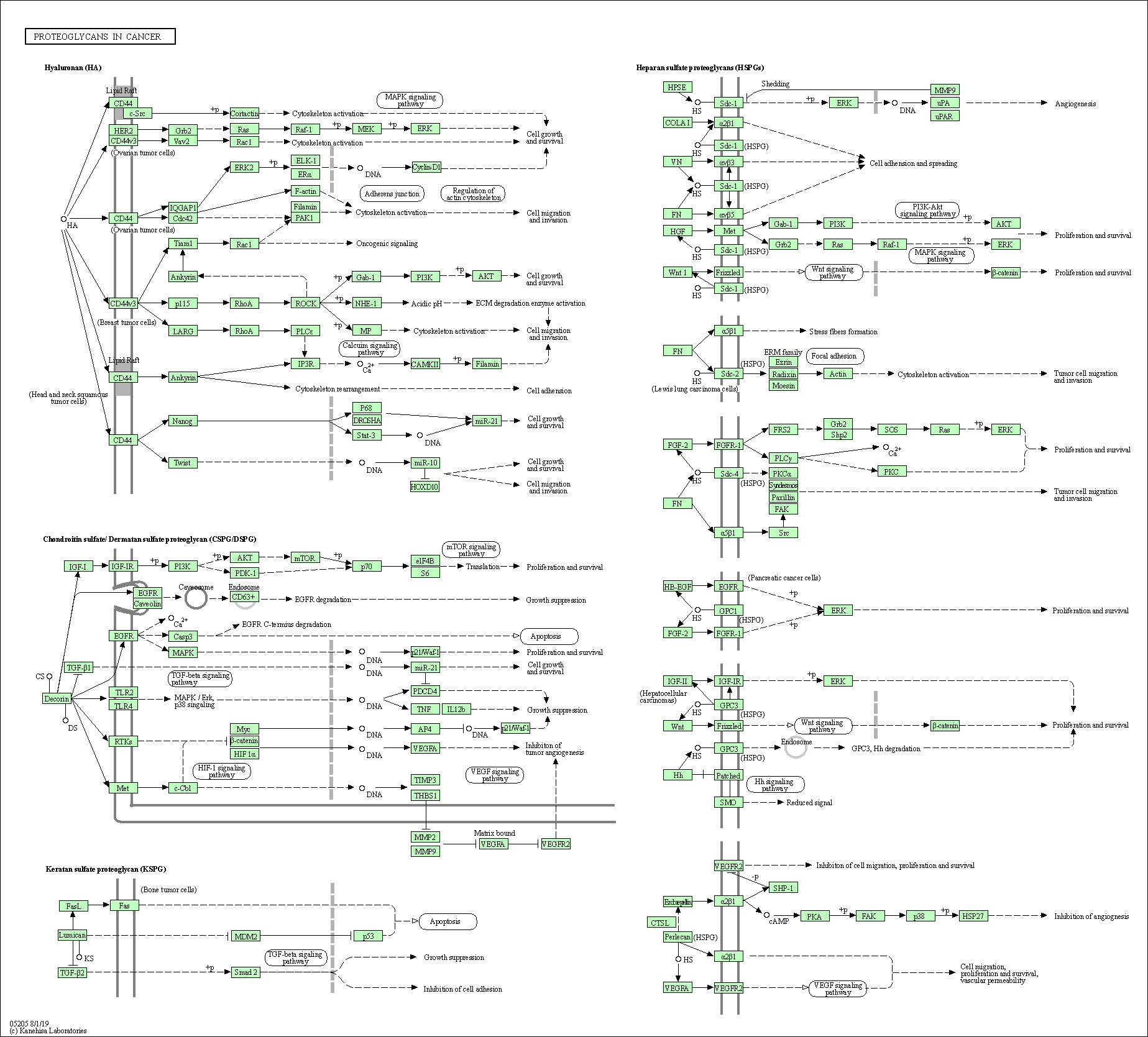
|
|||||||
| Hepatocellular carcinoma | hsa05225 |
Pathway Map 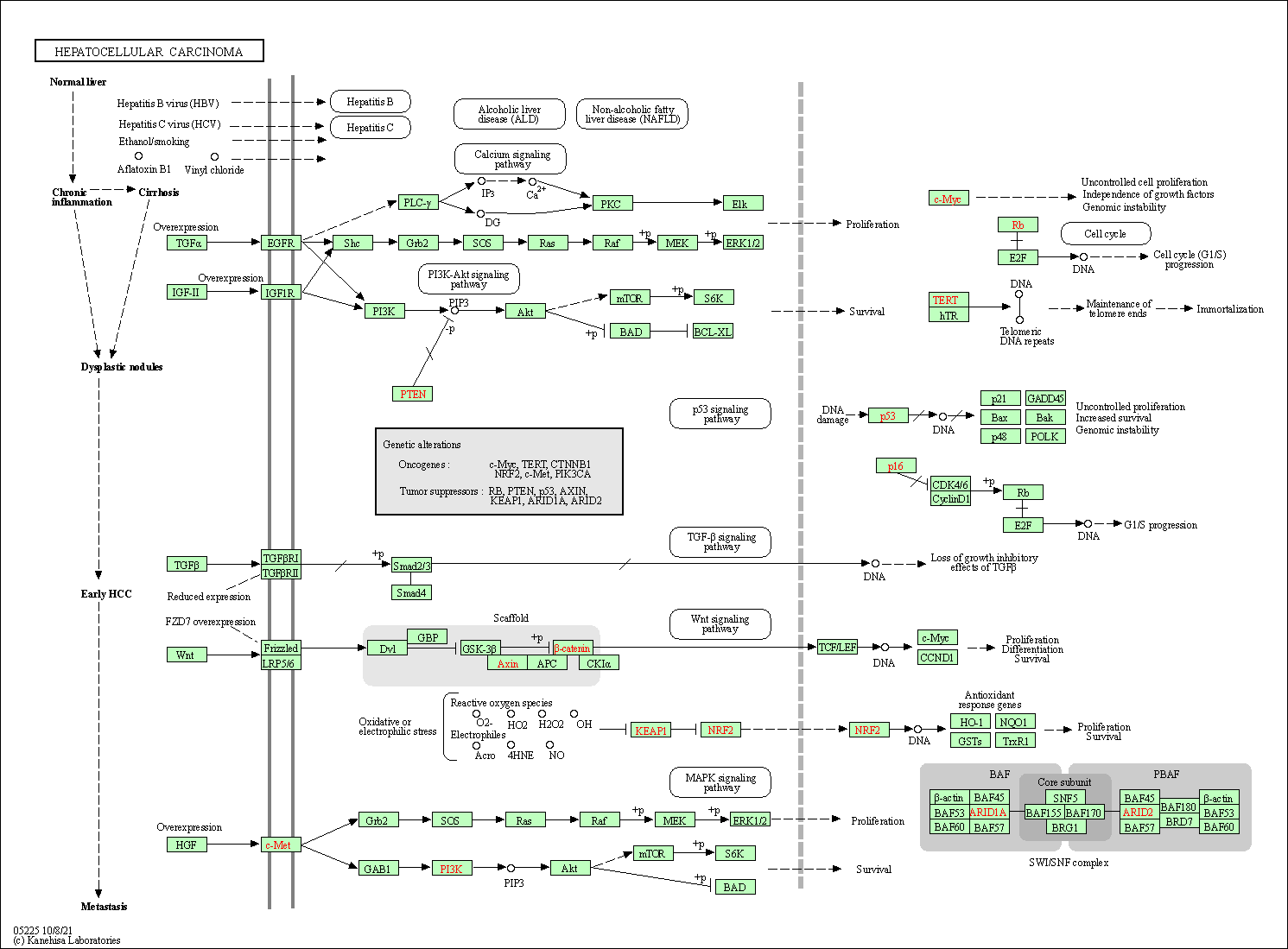
|
|||||||
| Hypertrophic cardiomyopathy | hsa05410 |
Pathway Map 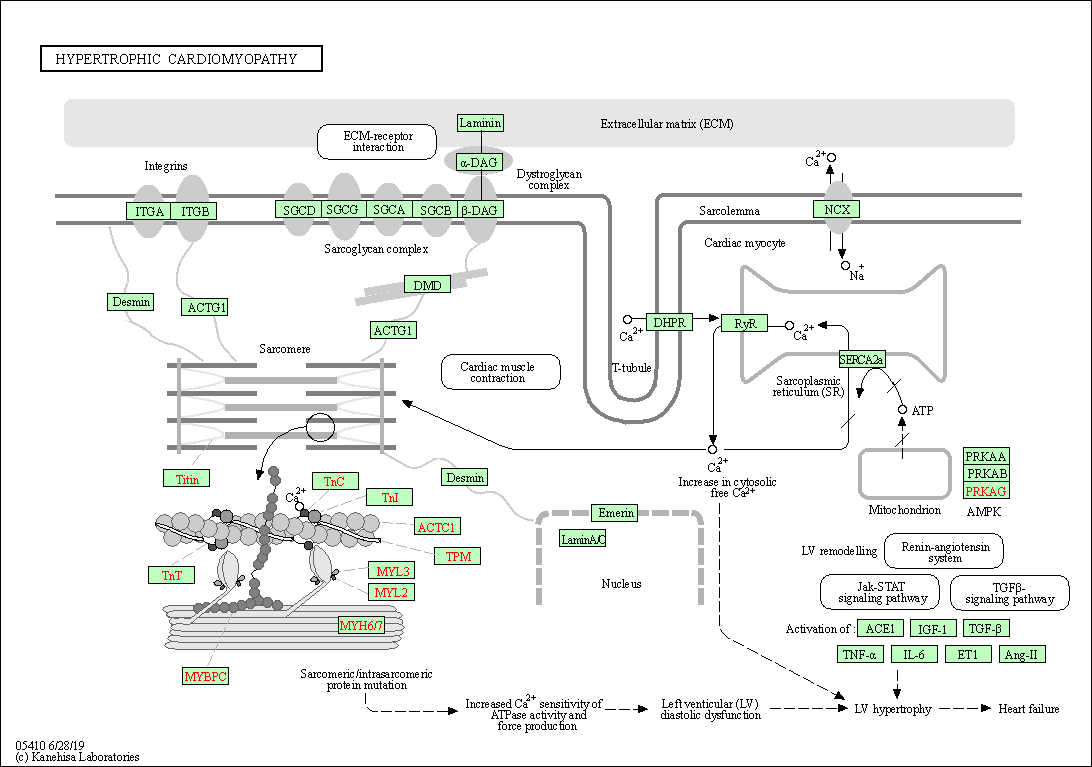
|
|||||||
| Arrhythmogenic right ventricular cardiomyopathy | hsa05412 |
Pathway Map 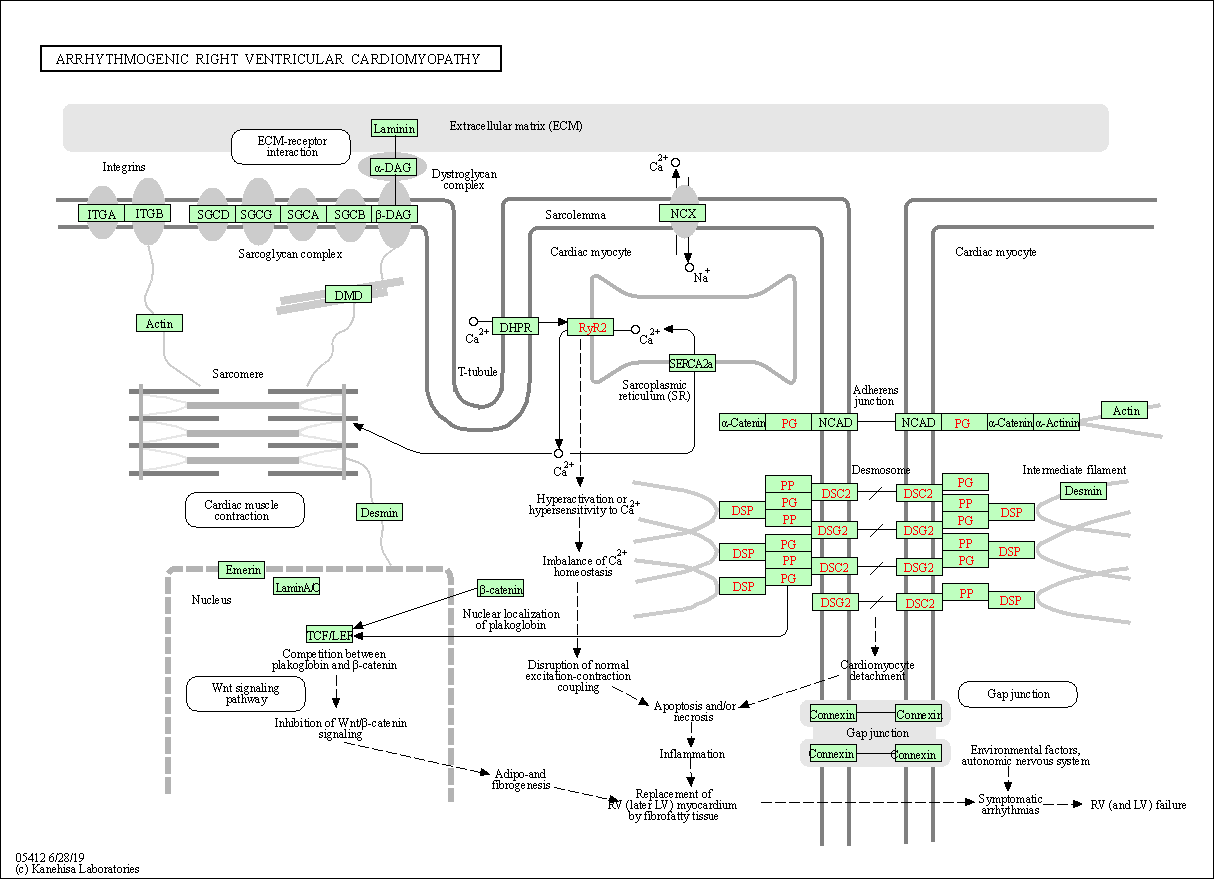
|
|||||||
| Dilated cardiomyopathy | hsa05414 |
Pathway Map 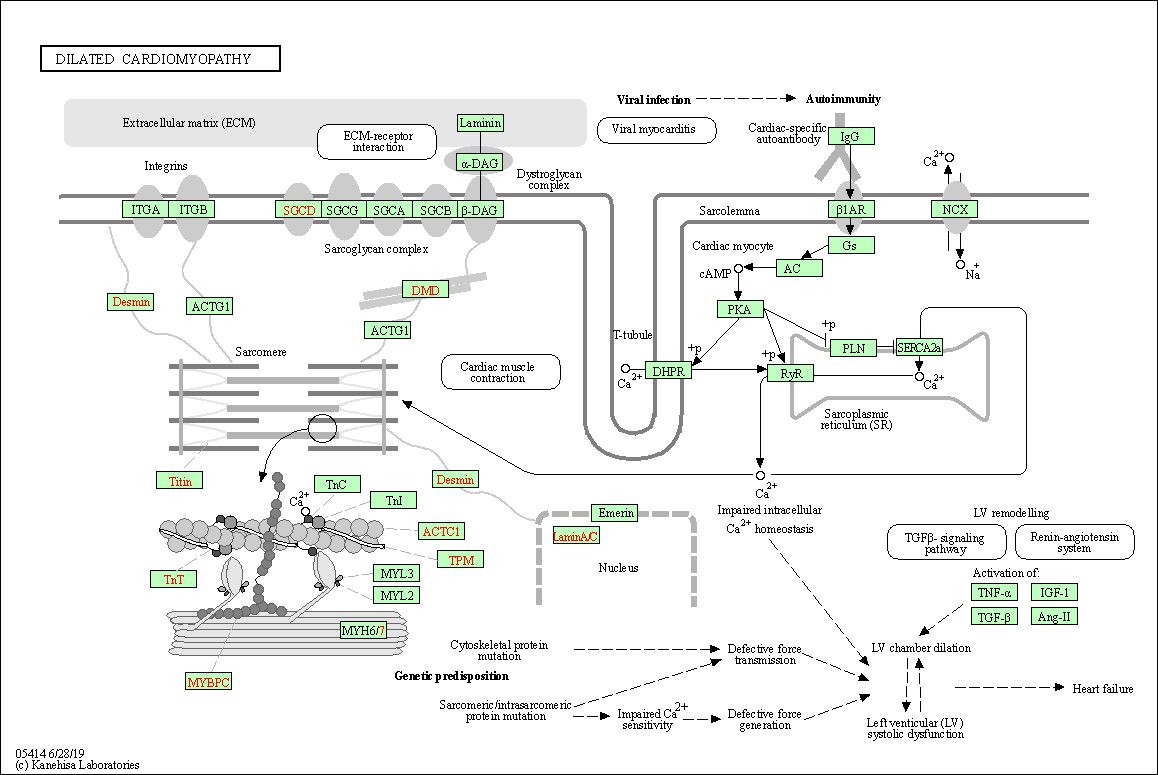
|
|||||||
| Viral myocarditis | hsa05416 |
Pathway Map 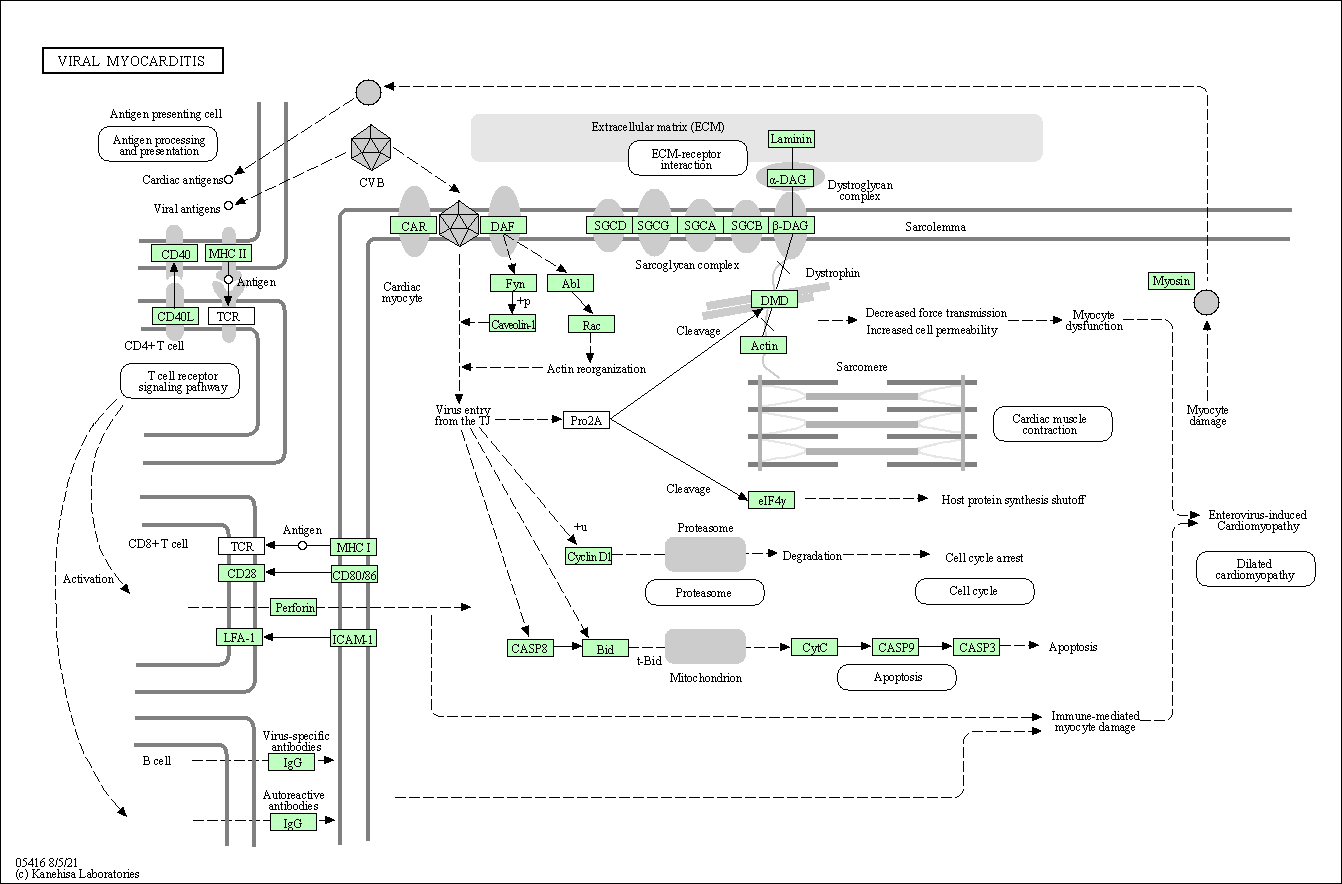
|
|||||||
| Fluid shear stress and atherosclerosis | hsa05418 |
Pathway Map 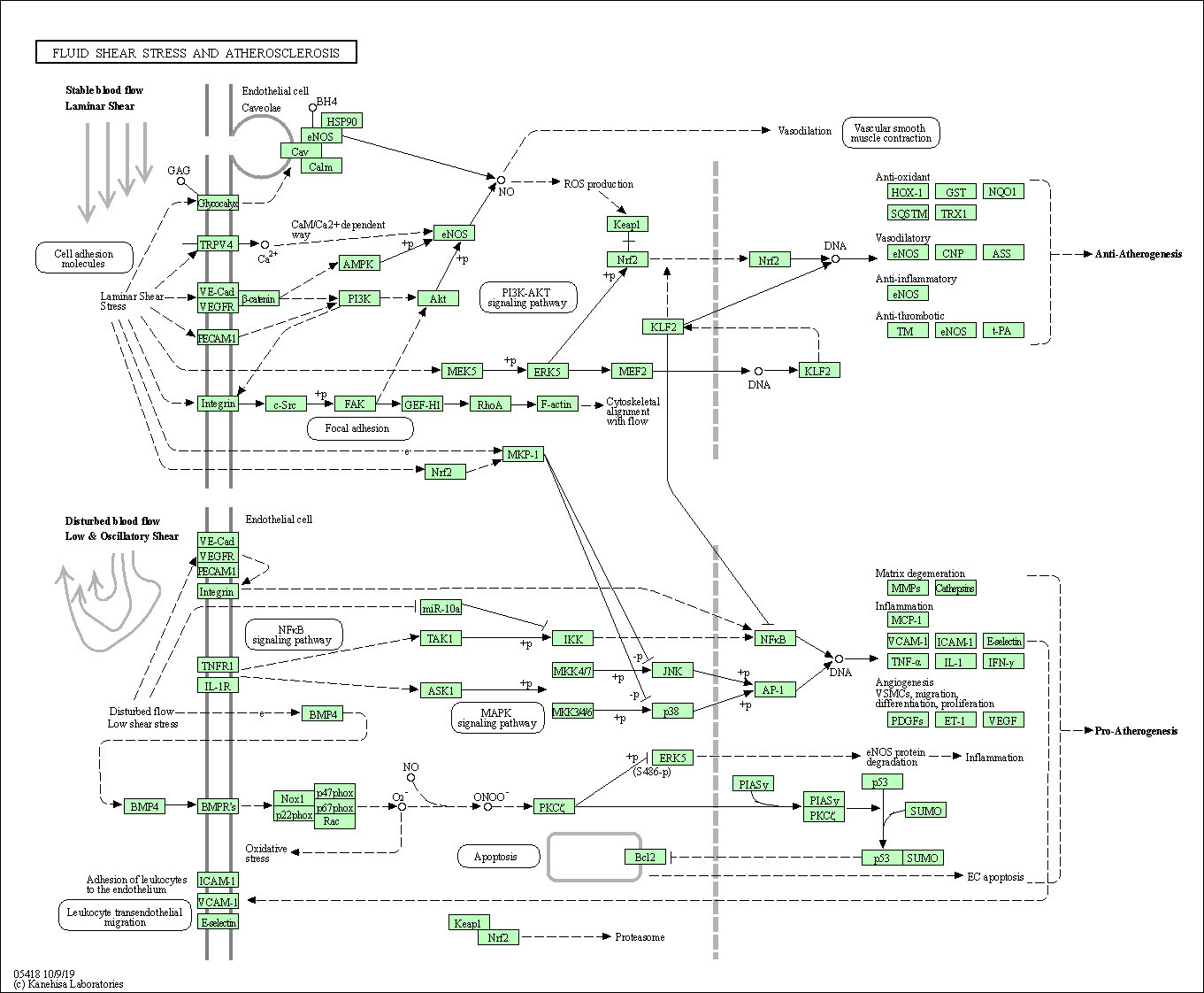
|
|||||||
| Apoptosis | hsa04210 |
Pathway Map 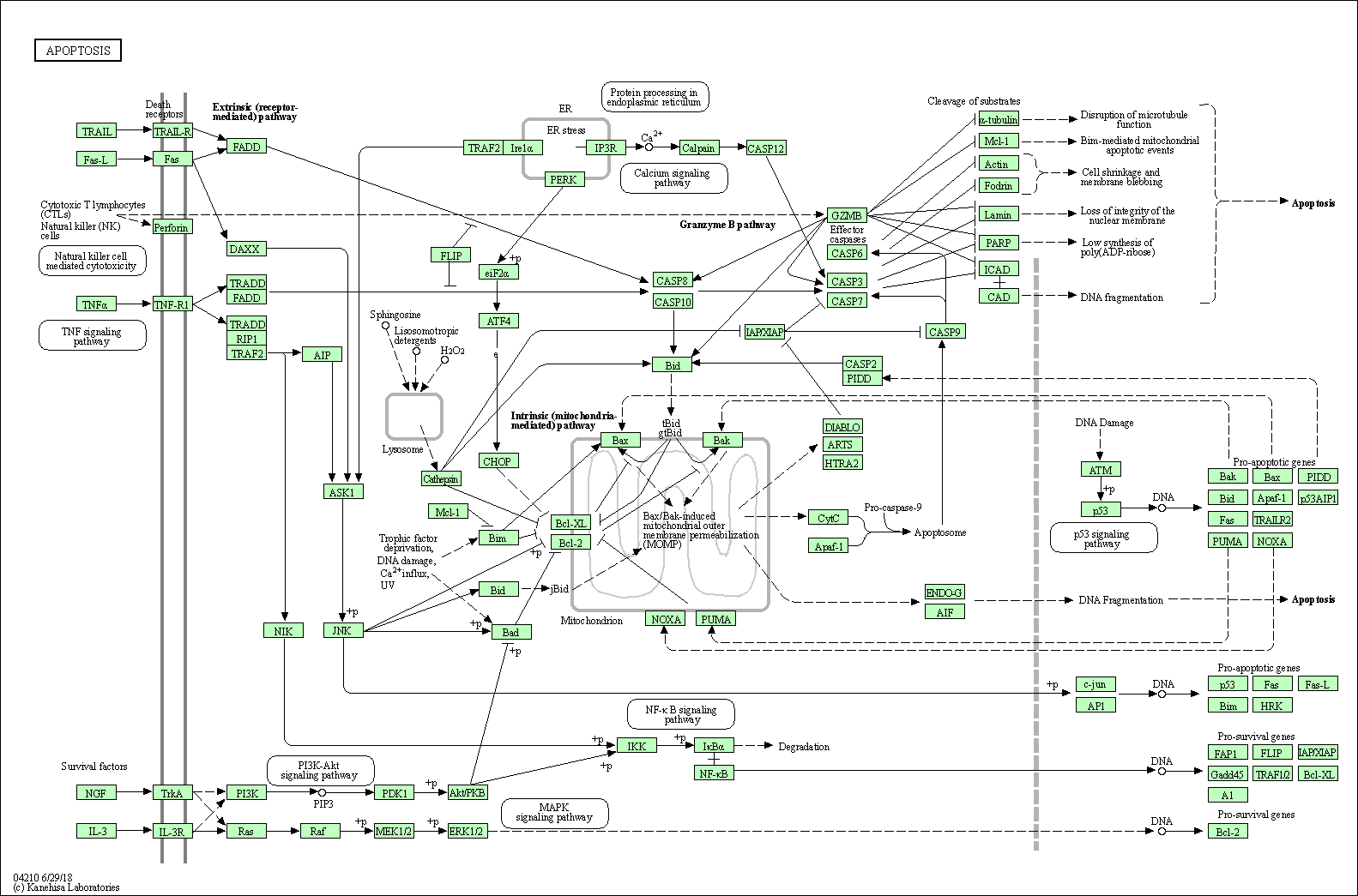
|
|||||||
| Regulation of actin cytoskeleton | hsa04810 |
Pathway Map 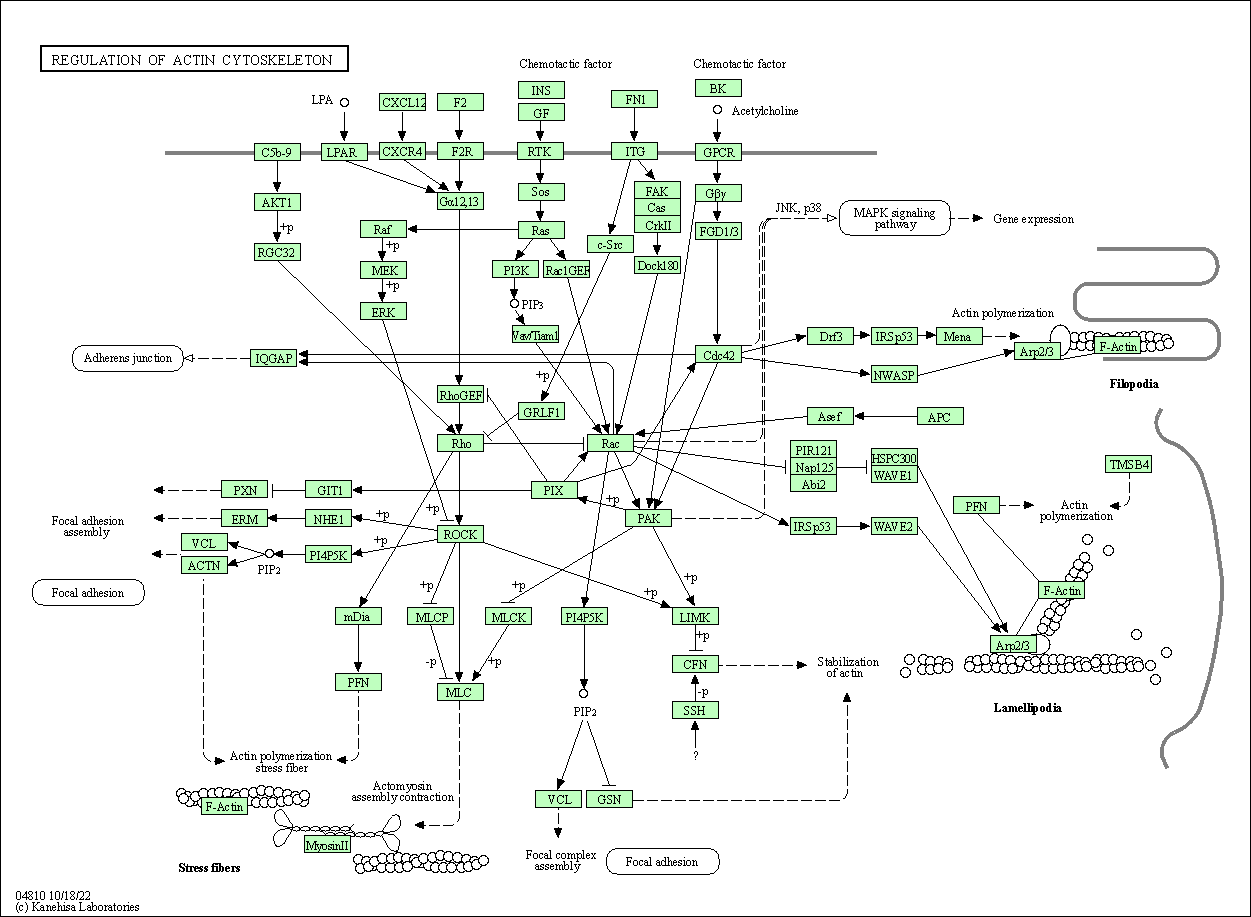
|
|||||||
| Motor proteins | hsa04814 |
Pathway Map 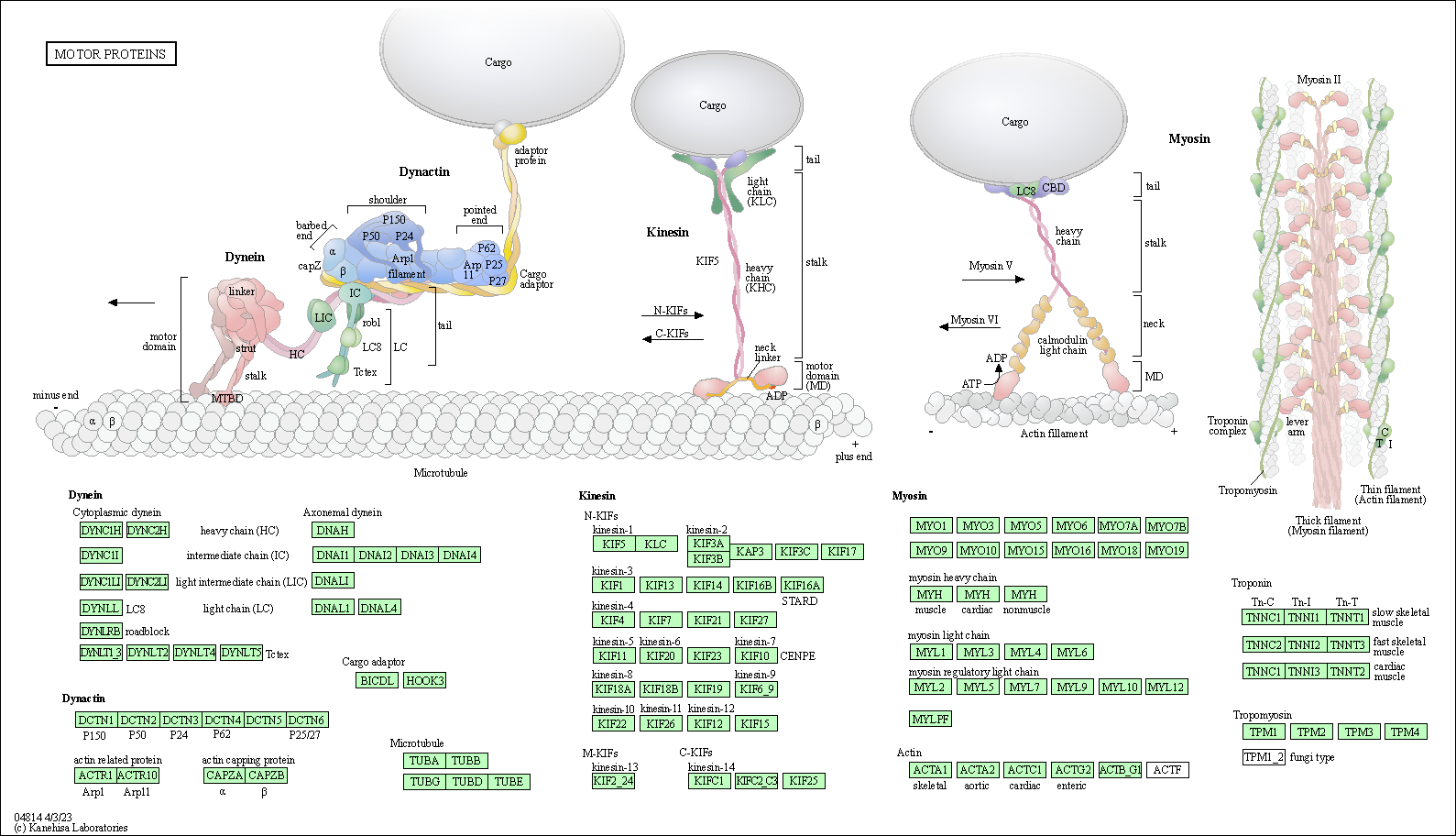
|
|||||||
| Focal adhesion | hsa04510 |
Pathway Map 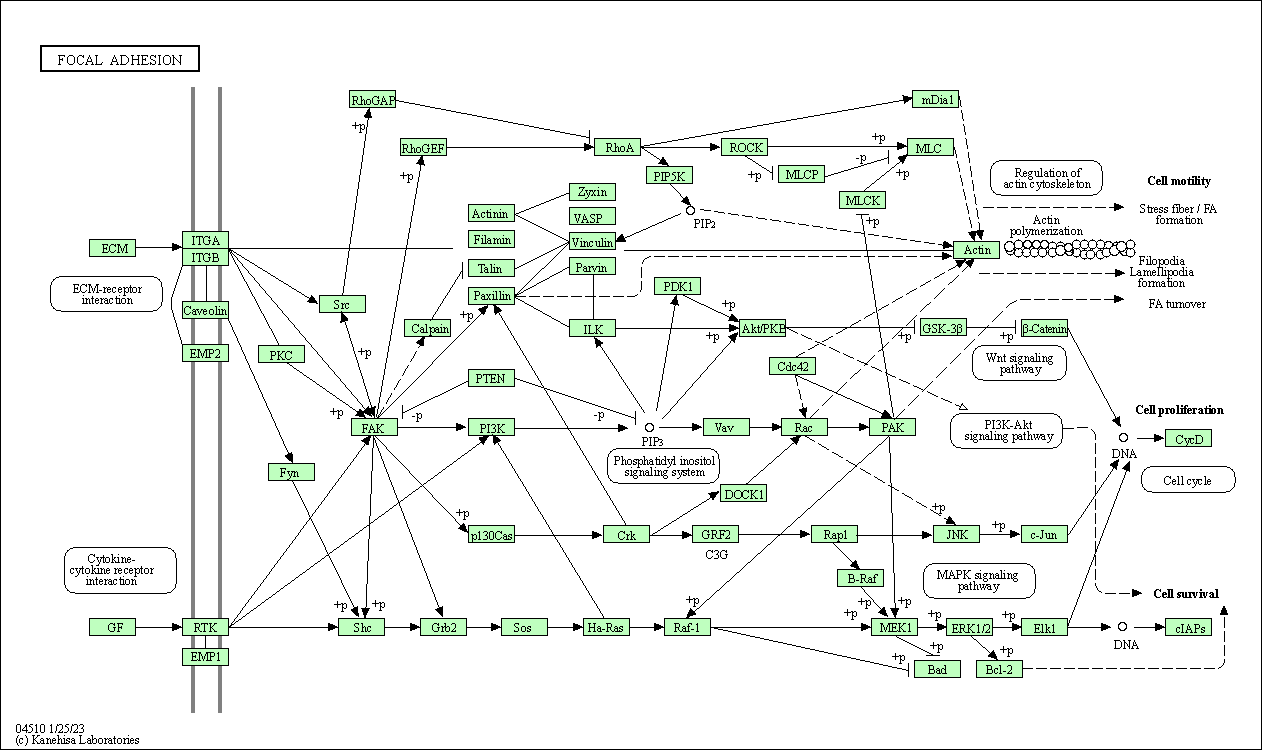
|
|||||||
| Adherens junction | hsa04520 |
Pathway Map 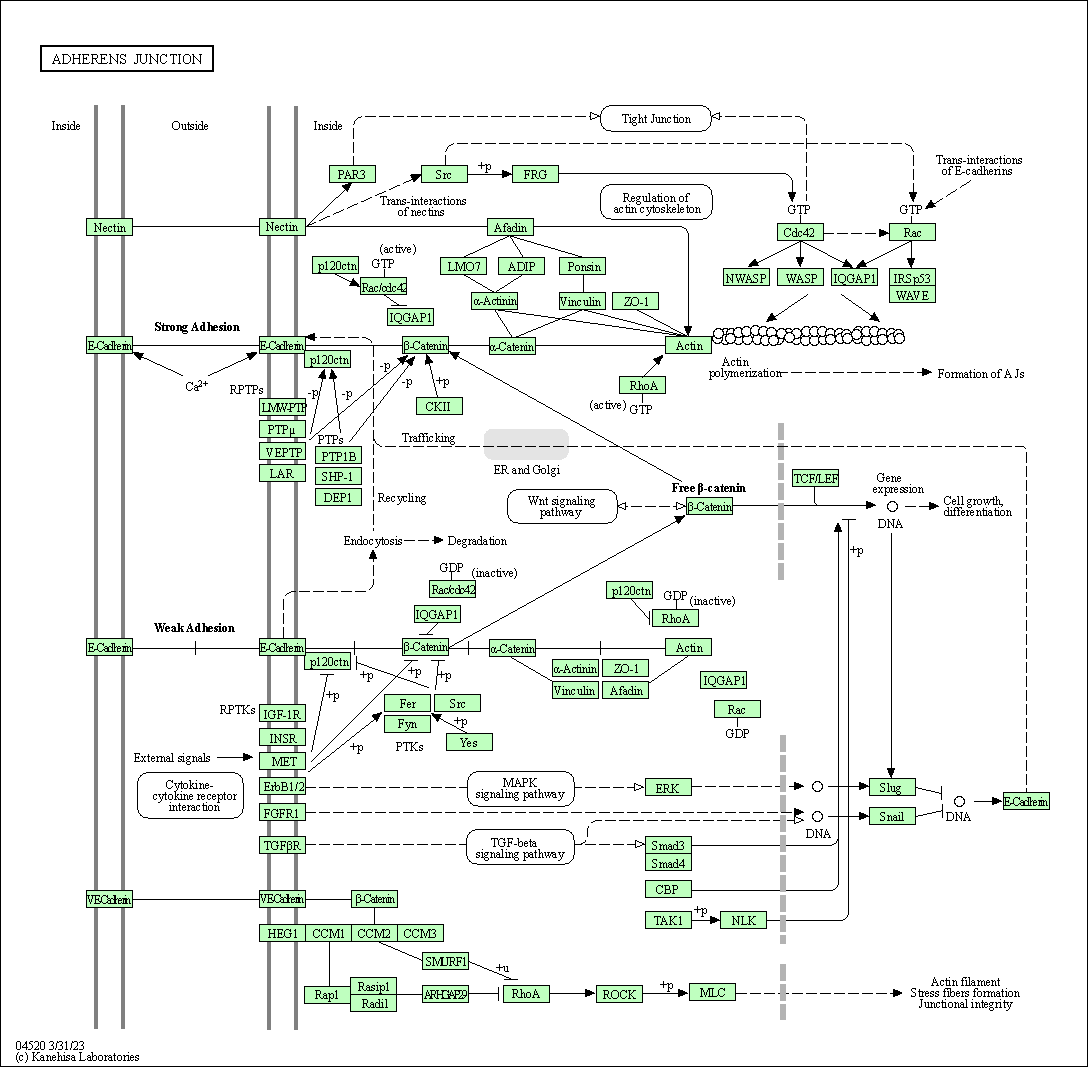
|
|||||||
| Tight junction | hsa04530 |
Pathway Map 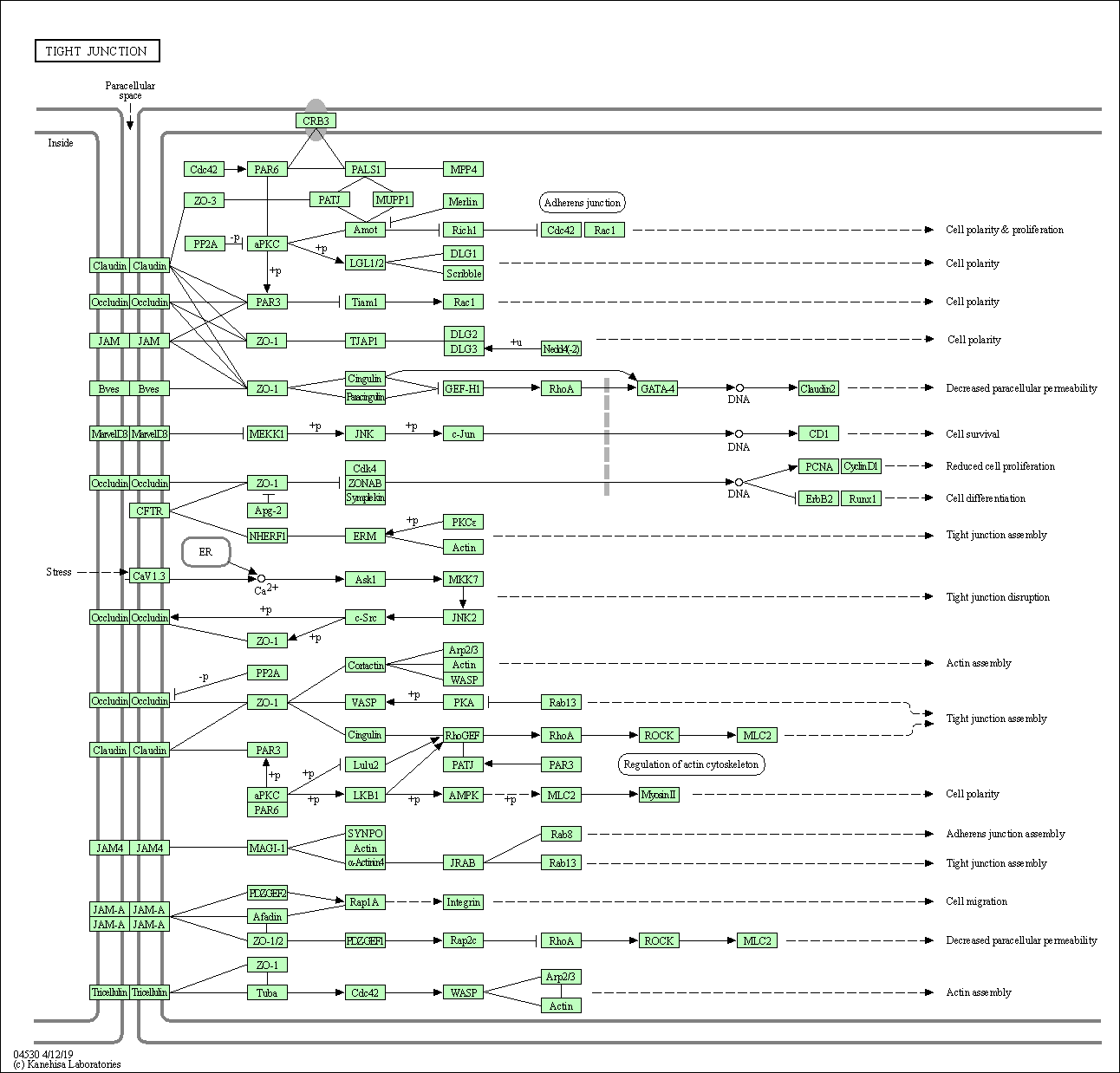
|
|||||||
| Gastric acid secretion | hsa04971 |
Pathway Map 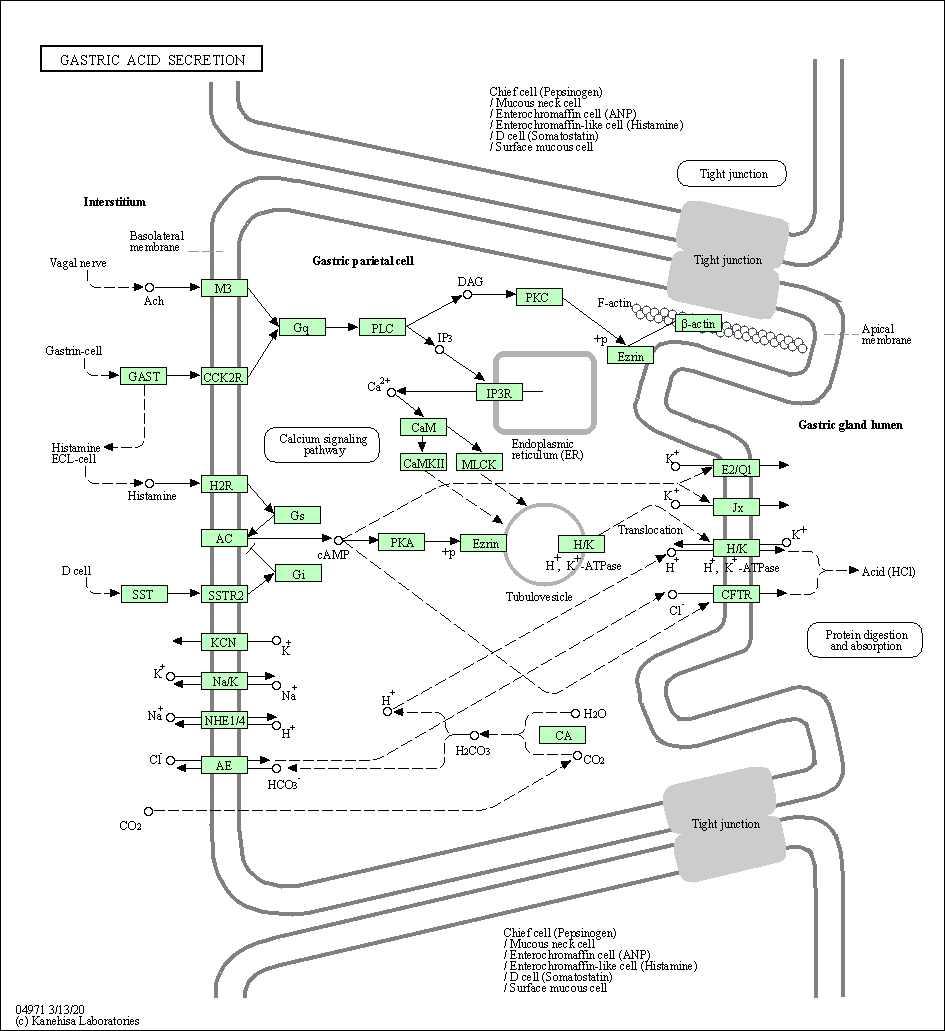
|
|||||||
| Thyroid hormone signaling pathway | hsa04919 |
Pathway Map 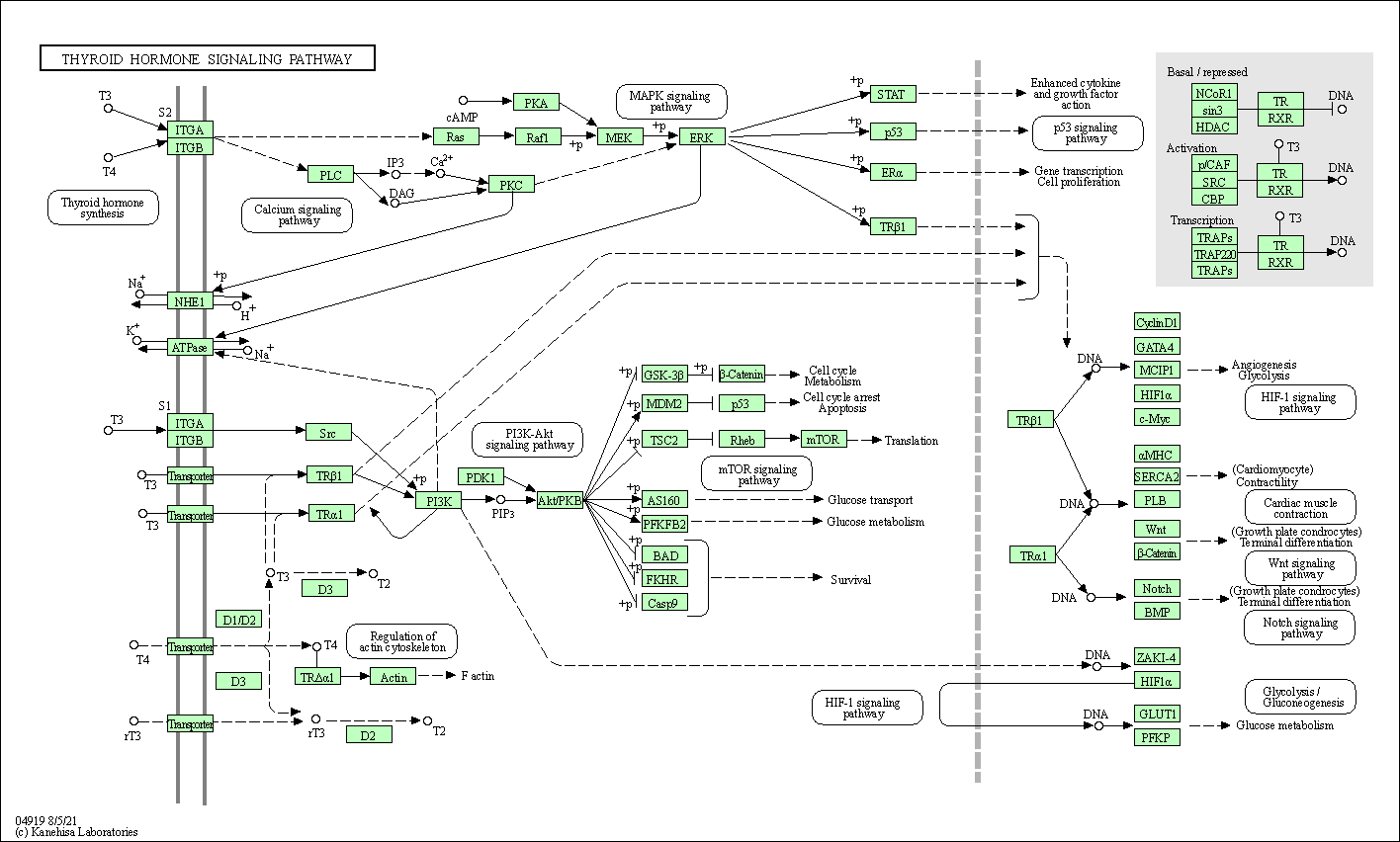
|
|||||||
| Oxytocin signaling pathway | hsa04921 |
Pathway Map 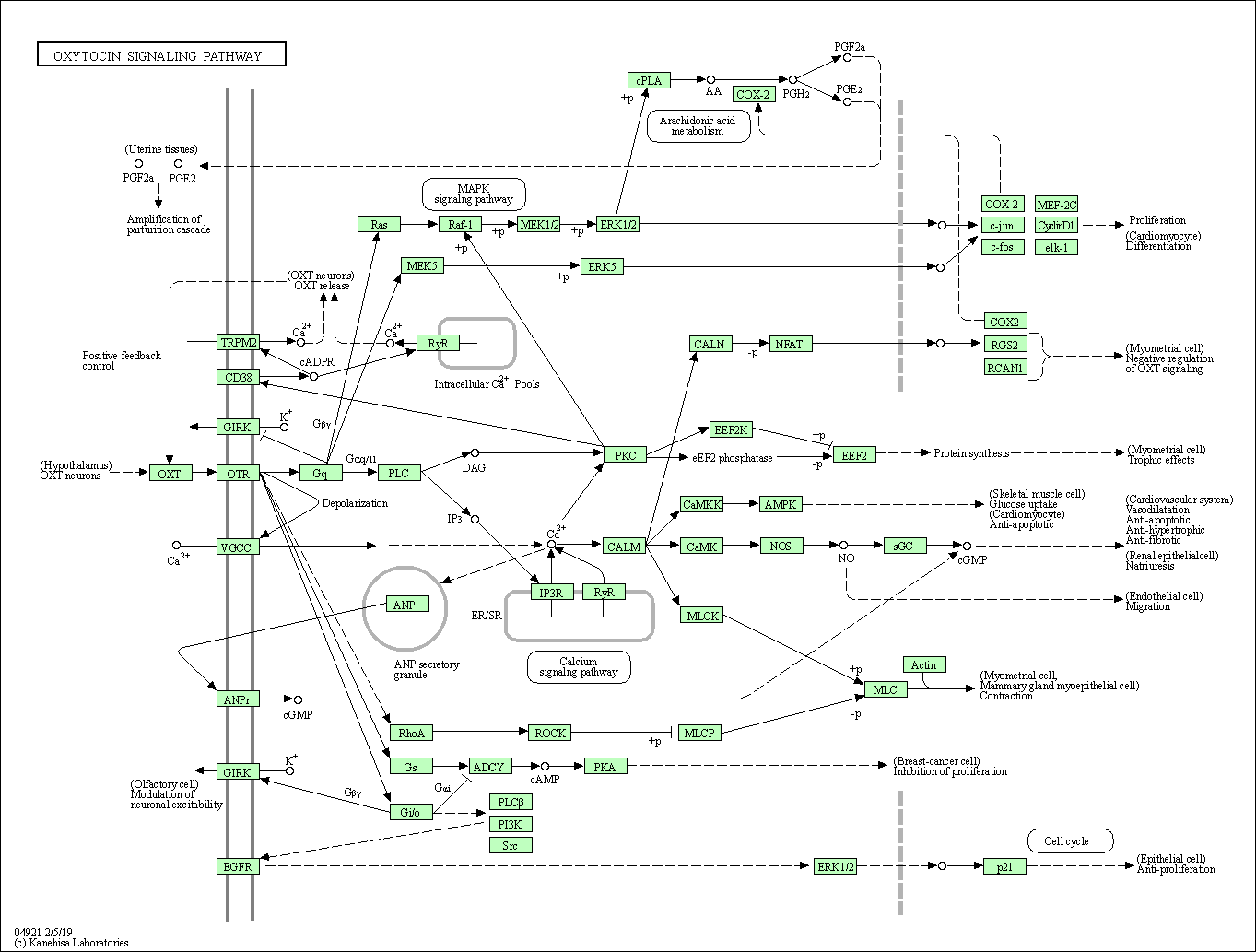
|
|||||||
| Thermogenesis | hsa04714 |
Pathway Map 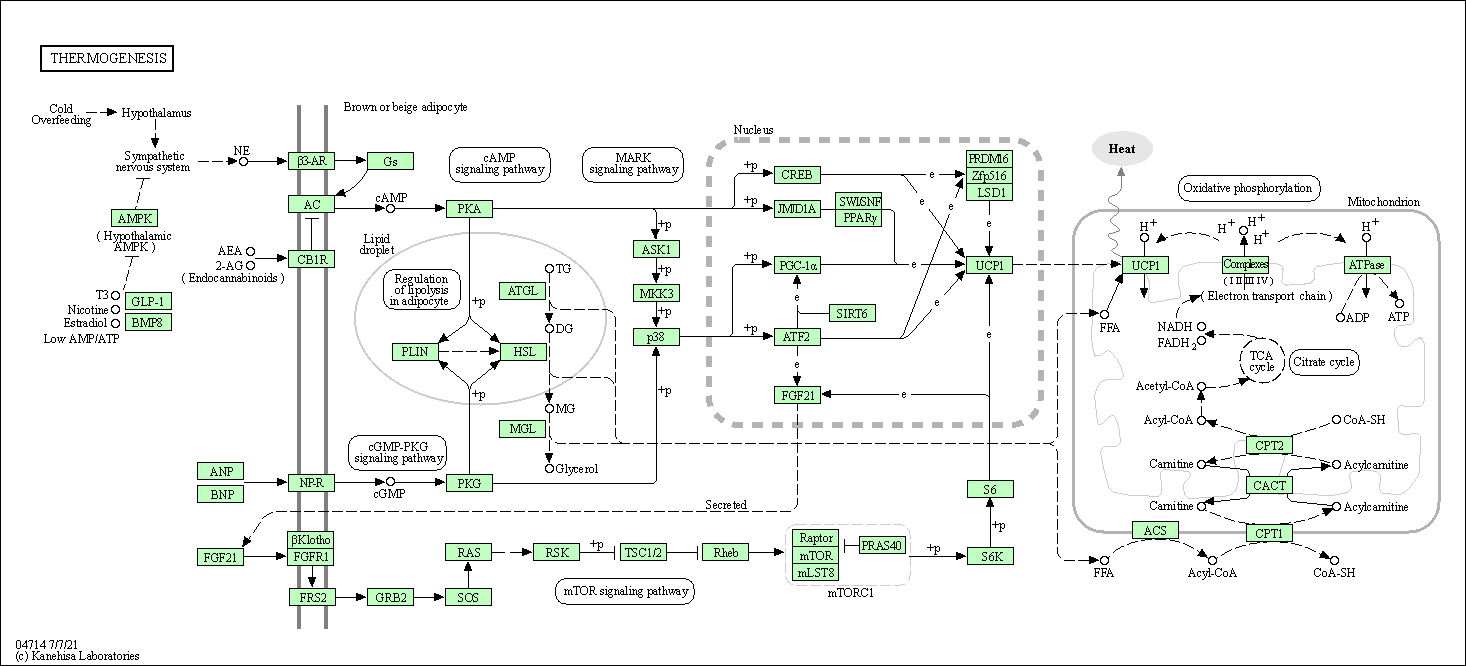
|
|||||||
| Amyotrophic lateral sclerosis | hsa05014 |
Pathway Map 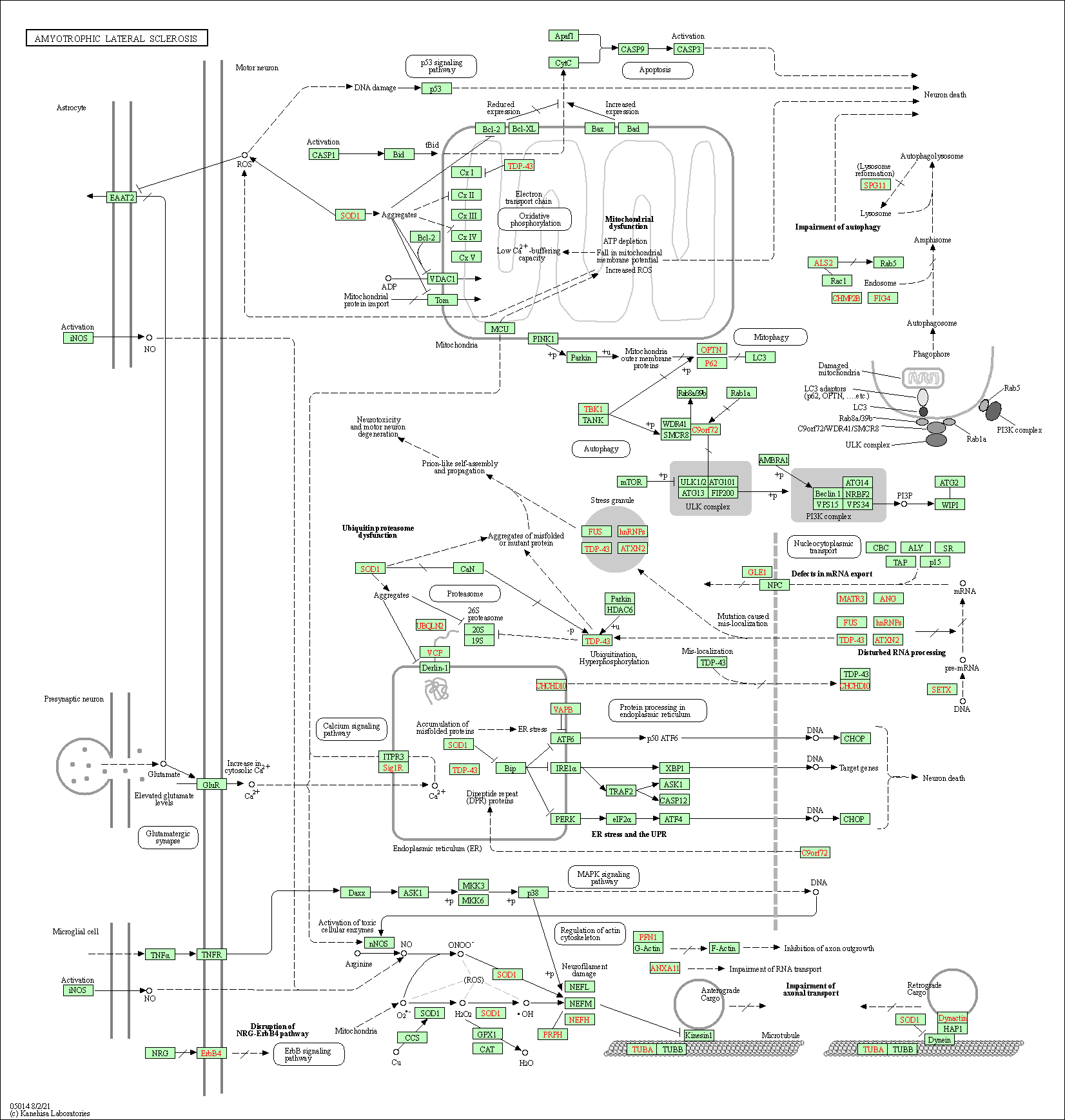
|
|||||||
| Rap1 signaling pathway | hsa04015 |
Pathway Map 
|
|||||||
| Hippo signaling pathway | hsa04390 |
Pathway Map 
|
|||||||
| Phagosome | hsa04145 |
Pathway Map 
|
|||||||
| 3D Structure |
|
||||||||
Full List of Virus RNA Interacting with This Protien
| Virus Name | Binding Region | RNA Info | Detection Method | Infection Cell | Cell ID | Cell Originated Tissue | Infection Time | Interaction Score 1 | Interaction Score 2 |
|---|---|---|---|---|---|---|---|---|---|
| Hepatitis C virus | 5'UTR | RNA Info | Matrix-assisted laser desorption/ionization mass spectrometry (MALDI-MS) | HuH-7 Cells (Human hepatocellular carcinoma cell) | . | Liver | . | . | . |
| Severe acute respiratory syndrome coronavirus 2 (strain hCoV-19/Not Specified Virus Strain) | Not Specified Virus Region | RNA Info | RNAantisensepurificationandquantitativemassspectrometry(RAP-MS); Tandemmasstag(TMT)labelling; liquidchromatographycoupledwithtandemmassspectrometry(LC-MS/MS); Westernblot | Huh7 cells (liver carcinoma celll ine) | . | Liver | 24h | log2 fold change = 4.71103E+14 | . |
| Severe acute respiratory syndrome coronavirus 2 (strain hCoV-19/IPBCAMS-YL01/2020) | 3'UTR | RNA Info | ChIRP-MS | Huh7.5.1 cells | . | Liver | 30 h | MIST = 0.691965934 | . |
Potential Drug(s) that Targets This Protein
| Drug Name | DrunkBank ID | Pubchem ID | TTD ID | REF | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CYCLOPHOSPHAMIDE | . | 2907 | D0CT9C | DGIdb | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| ETHINYL ESTRADIOL | . | 5991 | D06NXY | DGIdb | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Protein Sequence Information
|
MDDDIAALVVDNGSGMCKAGFAGDDAPRAVFPSIVGRPRHQGVMVGMGQKDSYVGDEAQSKRGILTLKYPIEHGIVTNWDDMEKIWHHTFYNELRVAPEEHPVLLTEAPLNPKANREKMTQIMFETFNTPAMYVAIQAVLSLYASGRTTGIVMDSGDGVTHTVPIYEGYALPHAILRLDLAGRDLTDYLMKILTERGYSFTTTAEREIVRDIKEKLCYVALDFEQEMATAASSSSLEKSYELPDGQVITIGNERFRCPEALFQPSFLGMESCGIHETTFNSIMKCDVDIRKDLYANTVLSGGTTMYPGIADRMQKEITALAPSTMKIKIIAPPERKYSVWIGGSILASLSTFQQMWISKQEYDESGPSIVHRKCF
Click to Show/Hide
|
