Host Protein General Information
| Protein Name |
Actin-binding protein WASF1
|
Gene Name |
WASF1
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Host Species |
Homo sapiens
|
Uniprot Entry Name |
WASF1_HUMAN
|
||||||
| Protein Families |
SCAR/WAVE family
|
||||||||
| Subcellular Location |
Cytoplasm; cytoskeleton
|
||||||||
| External Link | |||||||||
| NCBI Gene ID | |||||||||
| Uniprot ID | |||||||||
| Ensembl ID | |||||||||
| HGNC ID | |||||||||
| Related KEGG Pathway | |||||||||
| Fc gamma R-mediated phagocytosis | hsa04666 |
Pathway Map 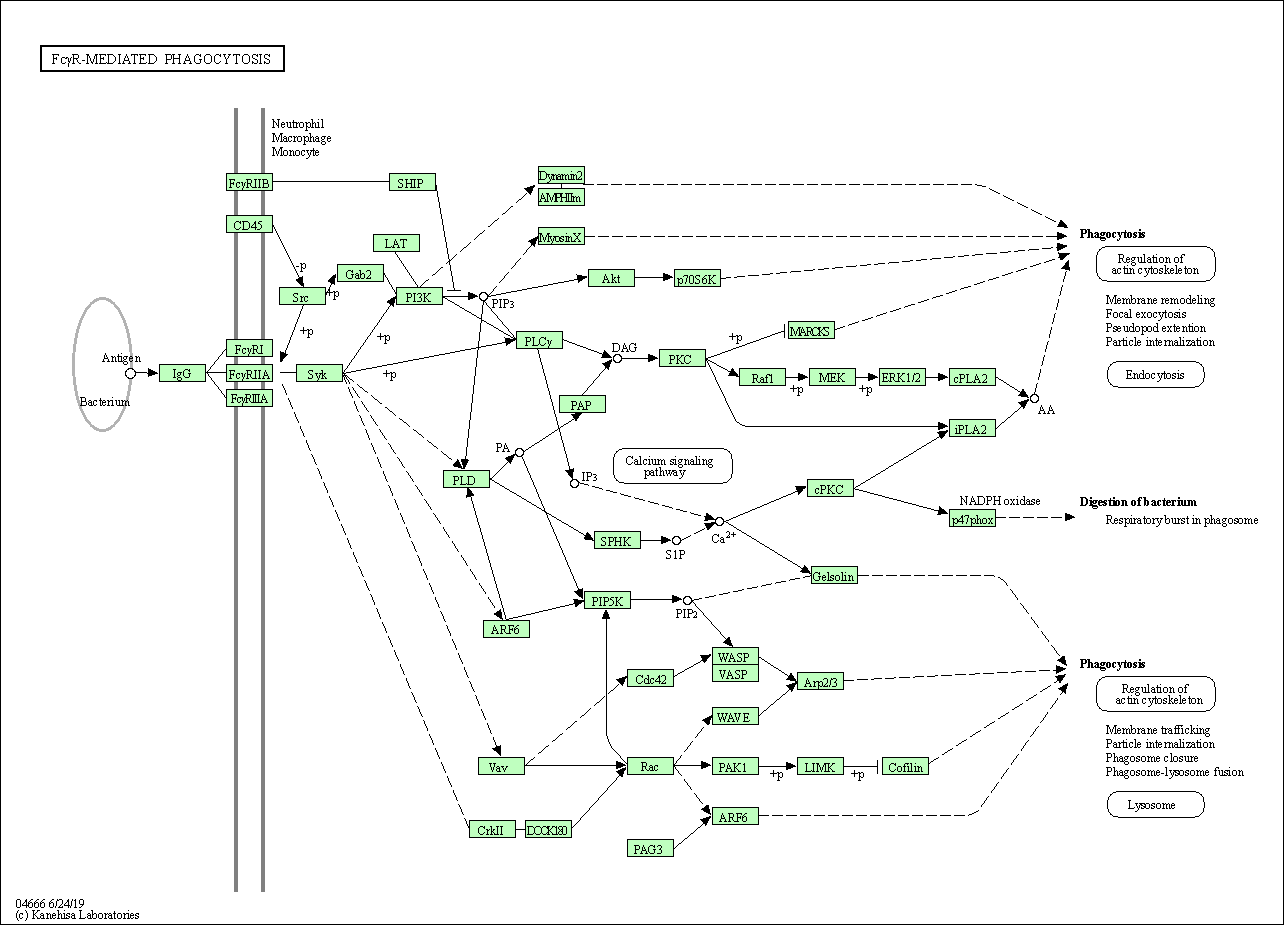
|
|||||||
| Bacterial invasion of epithelial cells | hsa05100 |
Pathway Map 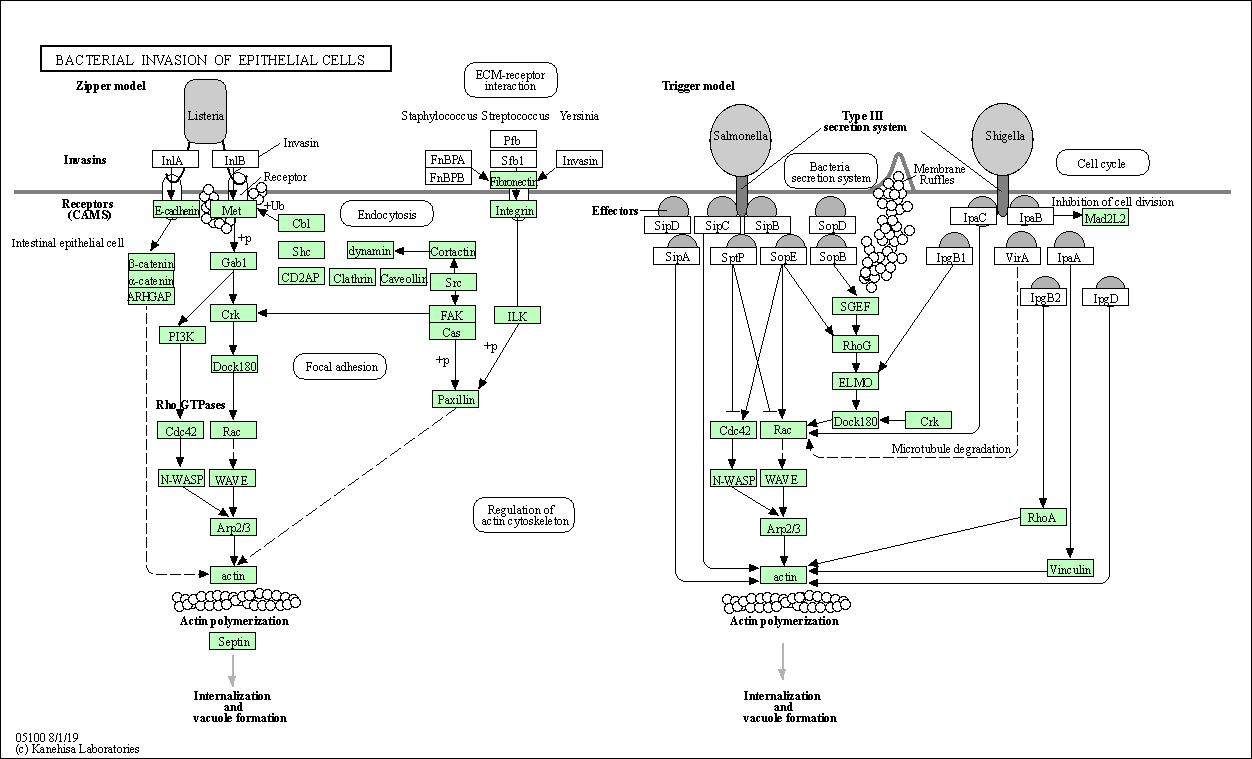
|
|||||||
| Pathogenic Escherichia coli infection | hsa05130 |
Pathway Map 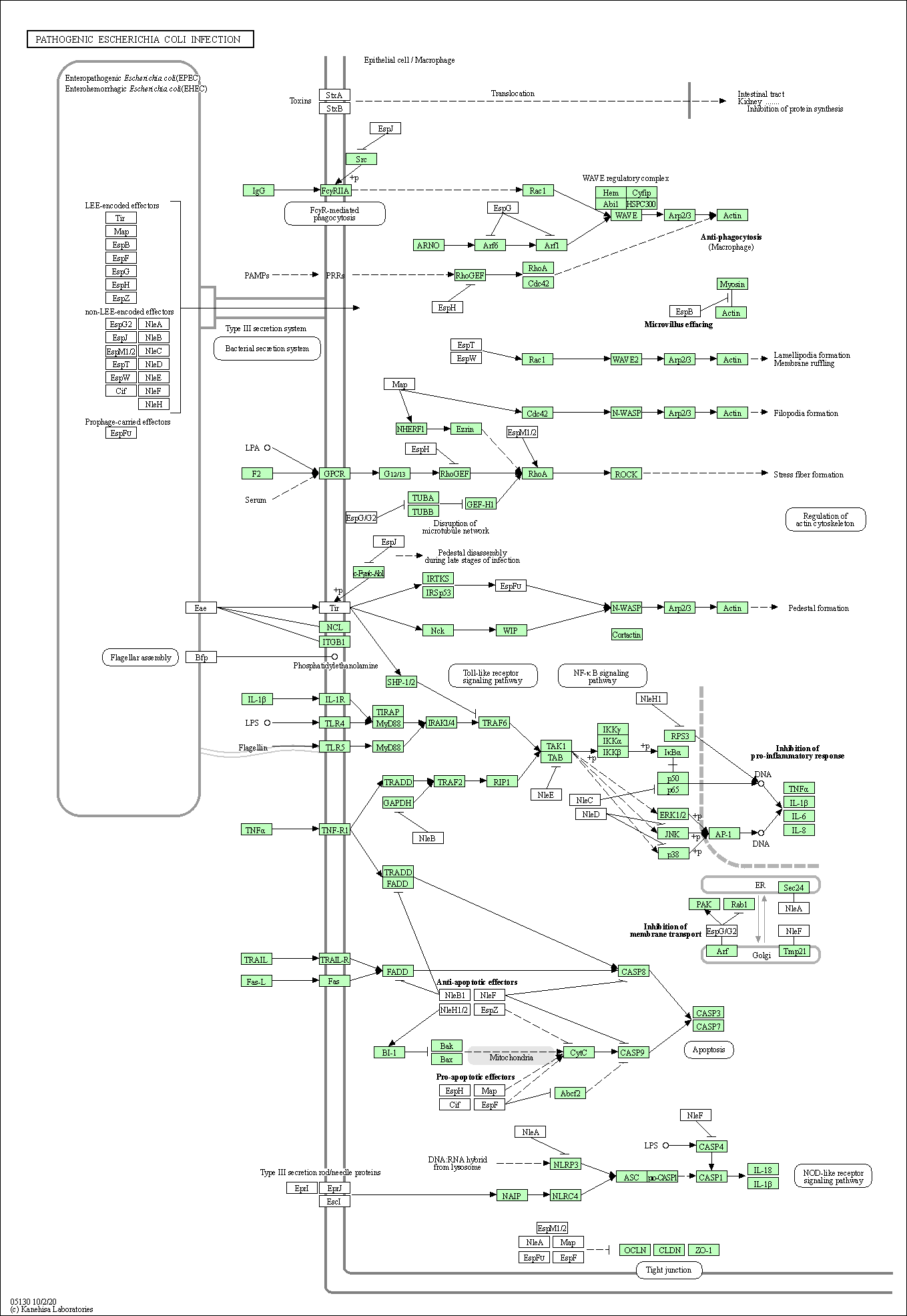
|
|||||||
| Shigellosis | hsa05131 |
Pathway Map 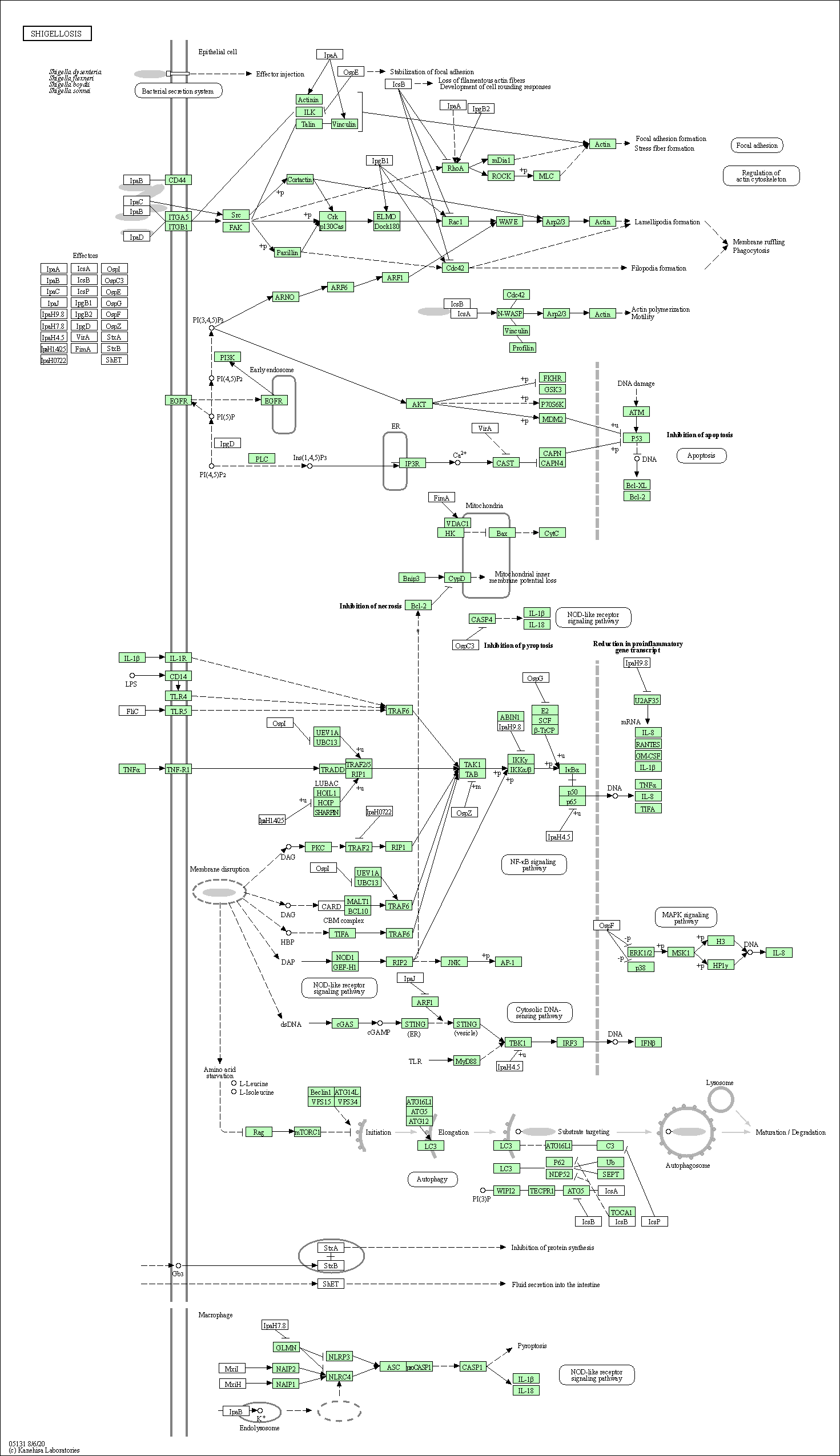
|
|||||||
| Choline metabolism in cancer | hsa05231 |
Pathway Map 
|
|||||||
| Regulation of actin cytoskeleton | hsa04810 |
Pathway Map 
|
|||||||
| Adherens junction | hsa04520 |
Pathway Map 
|
|||||||
| 3D Structure |
|
||||||||
Full List of Virus RNA Interacting with This Protien
| Virus Name | Binding Region | RNA Info | Detection Method | Infection Cell | Cell ID | Cell Originated Tissue | Infection Time | Interaction Score 1 | Interaction Score 2 |
|---|---|---|---|---|---|---|---|---|---|
| Dengue virus 2 (strain NGC) | 5'UTR - 3'UTR | RNA Info | VIRal Cross-Linking And Solid-phase Purification (VIR-CLASP) | HuH-7 Cells (Human hepatocellular carcinoma cell) | . | Liver | . | . | . |
Protein Sequence Information
|
MPLVKRNIDPRHLCHTALPRGIKNELECVTNISLANIIRQLSSLSKYAEDIFGELFNEAHSFSFRVNSLQERVDRLSVSVTQLDPKEEELSLQDITMRKAFRSSTIQDQQLFDRKTLPIPLQETYDVCEQPPPLNILTPYRDDGKEGLKFYTNPSYFFDLWKEKMLQDTEDKRKEKRKQKQKNLDRPHEPEKVPRAPHDRRREWQKLAQGPELAEDDANLLHKHIEVANGPASHFETRPQTYVDHMDGSYSLSALPFSQMSELLTRAEERVLVRPHEPPPPPPMHGAGDAKPIPTCISSATGLIENRPQSPATGRTPVFVSPTPPPPPPPLPSALSTSSLRASMTSTPPPPVPPPPPPPATALQAPAVPPPPAPLQIAPGVLHPAPPPIAPPLVQPSPPVARAAPVCETVPVHPLPQGEVQGLPPPPPPPPLPPPGIRPSSPVTVTALAHPPSGLHPTPSTAPGPHVPLMPPSPPSQVIPASEPKRHPSTLPVISDARSVLLEAIRKGIQLRKVEEQREQEAKHERIENDVATILSRRIAVEYSDSEDDSEFDEVDWLE
Click to Show/Hide
|
