Host Protein General Information
| Protein Name |
Sipa-1
|
Gene Name |
SIPA1
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Host Species |
Homo sapiens
|
Uniprot Entry Name |
SIPA1_HUMAN
|
||||||
| Subcellular Location |
Nucleus
|
||||||||
| External Link | |||||||||
| NCBI Gene ID | |||||||||
| Uniprot ID | |||||||||
| Ensembl ID | |||||||||
| HGNC ID | |||||||||
| Related KEGG Pathway | |||||||||
| Leukocyte transendothelial migration | hsa04670 |
Pathway Map 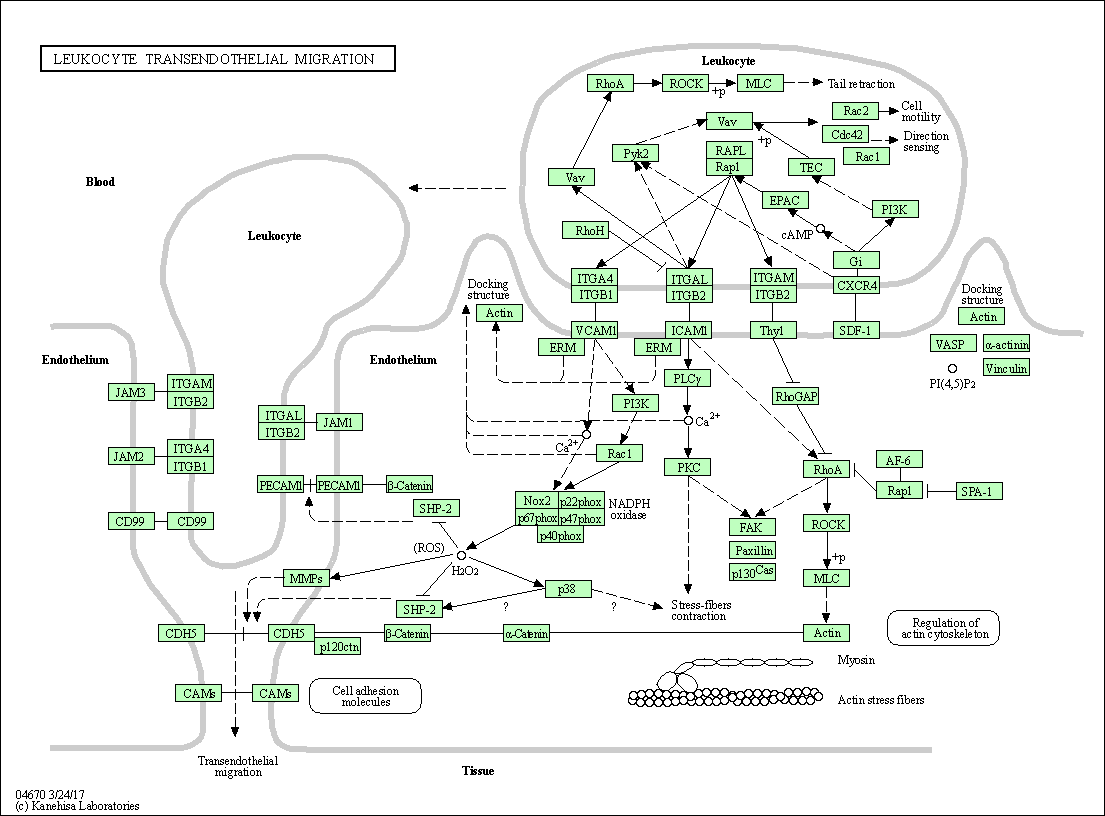
|
|||||||
| Rap1 signaling pathway | hsa04015 |
Pathway Map 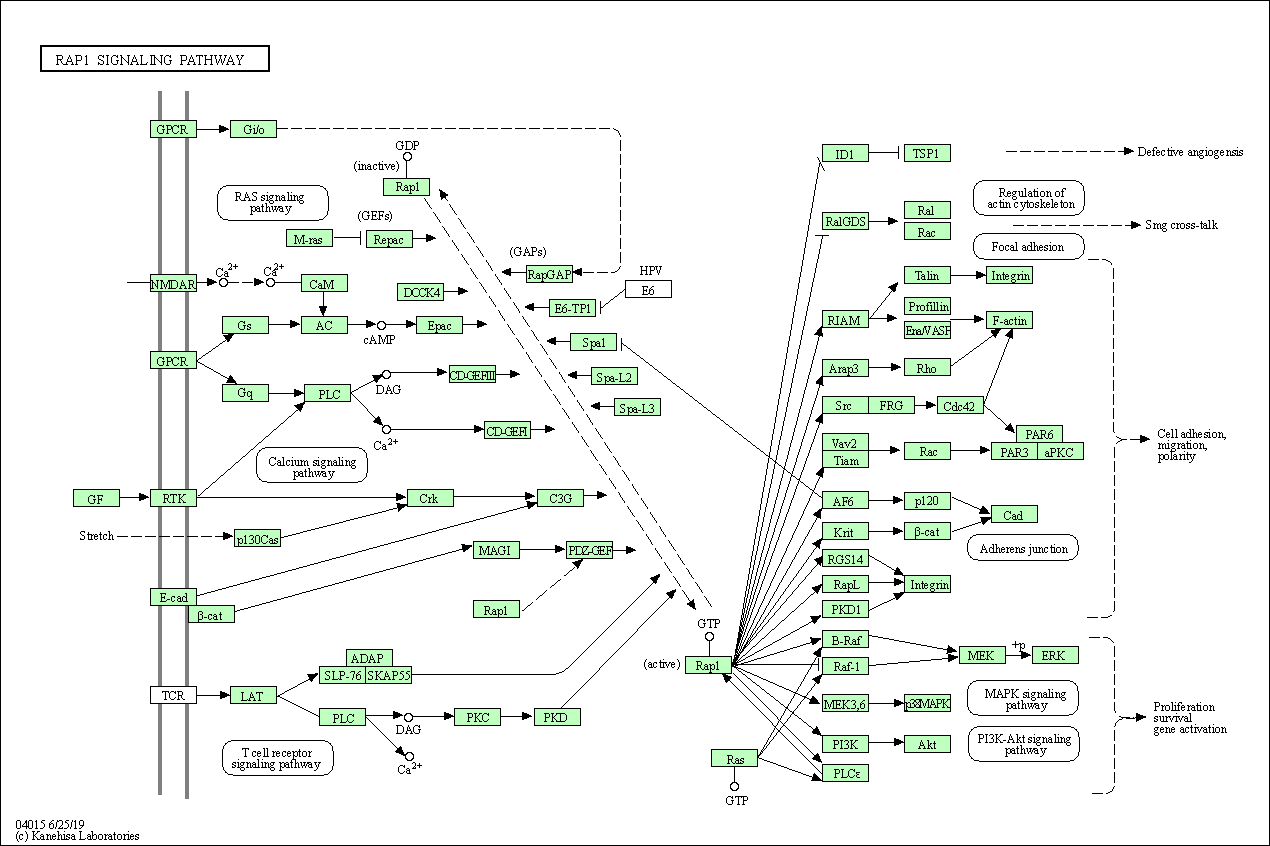
|
|||||||
| 3D Structure |
|
||||||||
Full List of Virus RNA Interacting with This Protien
| Virus Name | Binding Region | RNA Info | Detection Method | Infection Cell | Cell ID | Cell Originated Tissue | Infection Time | Interaction Score 1 | Interaction Score 2 |
|---|---|---|---|---|---|---|---|---|---|
| Chikungunya virus (strain 181/25) | 5'UTR - 3'UTR | RNA Info | VIRal Cross-Linking And Solid-phase Purification (VIR-CLASP) | BHK-21 Cells (Small hamster kidney fibroblast) | . | Kidney | 0.2 h | P = 0.0142765493426056 | . |
| Chikungunya virus (strain 181/25) | 5'UTR - 3'UTR | RNA Info | VIRal Cross-Linking And Solid-phase Purification (VIR-CLASP) | BHK-21 Cells (Small hamster kidney fibroblast) | . | Kidney | 1 h | P = 0.0023856583461 | . |
| Chikungunya virus (strain 181/25) | 5'UTR - 3'UTR | RNA Info | VIRal Cross-Linking And Solid-phase Purification (VIR-CLASP) | BHK-21 Cells (Small hamster kidney fibroblast) | . | Kidney | 3 h | P = 0.000109019844078903 | . |
Protein Sequence Information
|
MPMWAGGVGSPRRGMAPASTDDLFARKLRQPARPPLTPHTFEPRPVRGPLLRSGSDAGEARPPTPASPRARAHSHEEASRPAATSTRLFTDPLALLGLPAEEPEPAFPPVLEPRWFAHYDVQSLLFDWAPRSQGMGSHSEASSGTLASAEDQAASSDLLHGAPGFVCELGGEGELGLGGPASPPVPPALPNAAVSILEEPQNRTSAYSLEHADLGAGYYRKYFYGKEHQNFFGMDESLGPVAVSLRREEKEGSGGGTLHSYRVIVRTTQLRTLRGTISEDALPPGPPRGLSPRKLLEHVAPQLSPSCLRLGSASPKVPRTLLTLDEQVLSFQRKVGILYCRAGQGSEEEMYNNQEAGPAFMQFLTLLGDVVRLKGFESYRAQLDTKTDSTGTHSLYTTYQDHEIMFHVSTMLPYTPNNQQQLLRKRHIGNDIVTIVFQEPGSKPFCPTTIRSHFQHVFLVVRAHTPCTPHTTYRVAVSRTQDTPAFGPALPAGGGPFAANADFRAFLLAKALNGEQAAGHARQFHAMATRTRQQYLQDLATNEVTTTSLDSASRFGLPSLGGRRRAAPRGPGAELQAAGSLVWGVRAAPGARVAAGAQASGPEGIEVPCLLGISAEALVLVAPRDGRVVFNCACRDVLAWTFSEQQLDLYHGRGEAITLRFDGSPGQAVGEVVARLQLVSRGCETRELALPRDGQGRLGFEVDAEGFVTHVERFTFAETAGLRPGARLLRVCGQTLPSLRPEAAAQLLRSAPKVCVTVLPPDESGRPRRSFSELYTLSLQEPSRRGAPDPVQDEVQGVTLLPTTKQLLHLCLQDGGSPPGPGDLAEERTEFLHSQNSLSPRSSLSDEAPVLPNTTPDLLLATTAKPSVPSADSETPLTQDRPGSPSGSEDKGNPAPELRASFLPRTLSLRNSISRIMSEAGSGTLEDEWQAISEIASTCNTILESLSREGQPIPESGDPKGTPKSDAEPEPGNLSEKVSHLESMLRKLQEDLQKEKADRAALEEEVRSLRHNNRRLQAESESAATRLLLASKQLGSPTADLA
Click to Show/Hide
|
