Host Protein General Information
| Protein Name |
PKA C-beta
|
Gene Name |
PRKACB
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Host Species |
Homo sapiens
|
Uniprot Entry Name |
KAPCB_HUMAN
|
||||||
| Protein Families |
Protein kinase superfamily, AGC Ser/Thr protein kinase family, cAMP subfamily
|
||||||||
| EC Number |
2.7.11.11
|
||||||||
| Subcellular Location |
Cytoplasm
|
||||||||
| External Link | |||||||||
| NCBI Gene ID | |||||||||
| Uniprot ID | |||||||||
| Ensembl ID | |||||||||
| HGNC ID | |||||||||
| Related KEGG Pathway | |||||||||
| Chemokine signaling pathway | hsa04062 |
Pathway Map 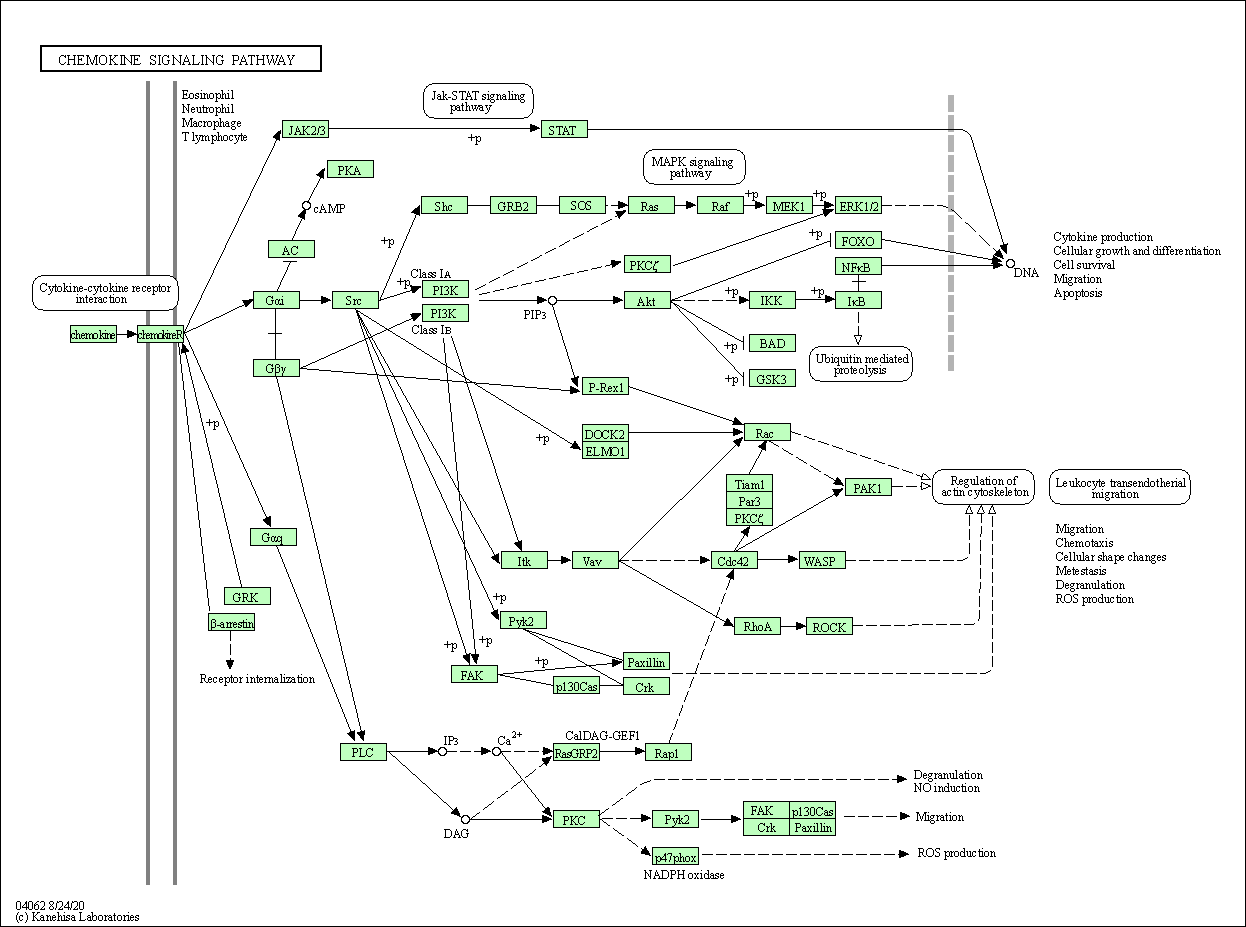
|
|||||||
| Platelet activation | hsa04611 |
Pathway Map 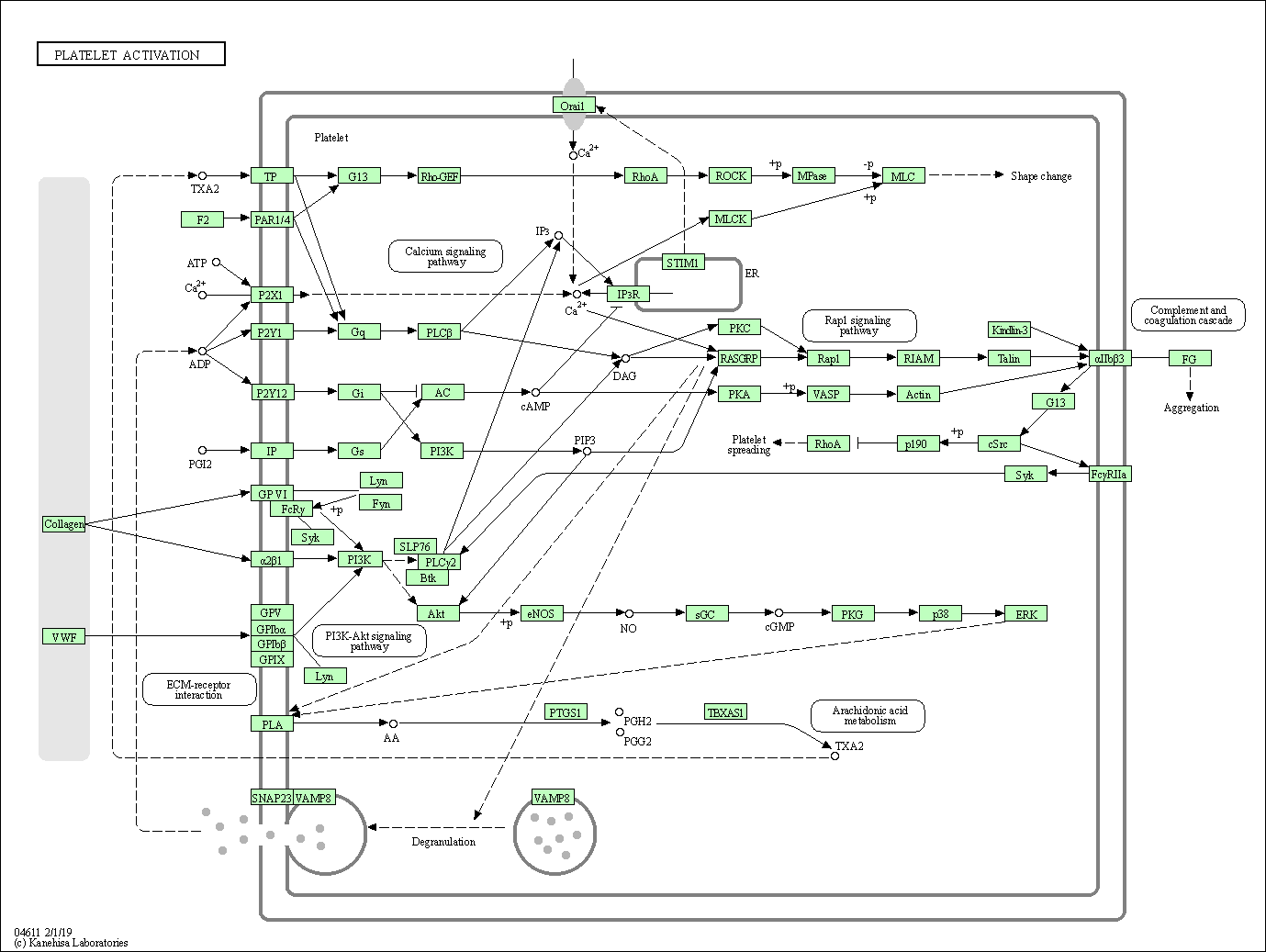
|
|||||||
| Vibrio cholerae infection | hsa05110 |
Pathway Map 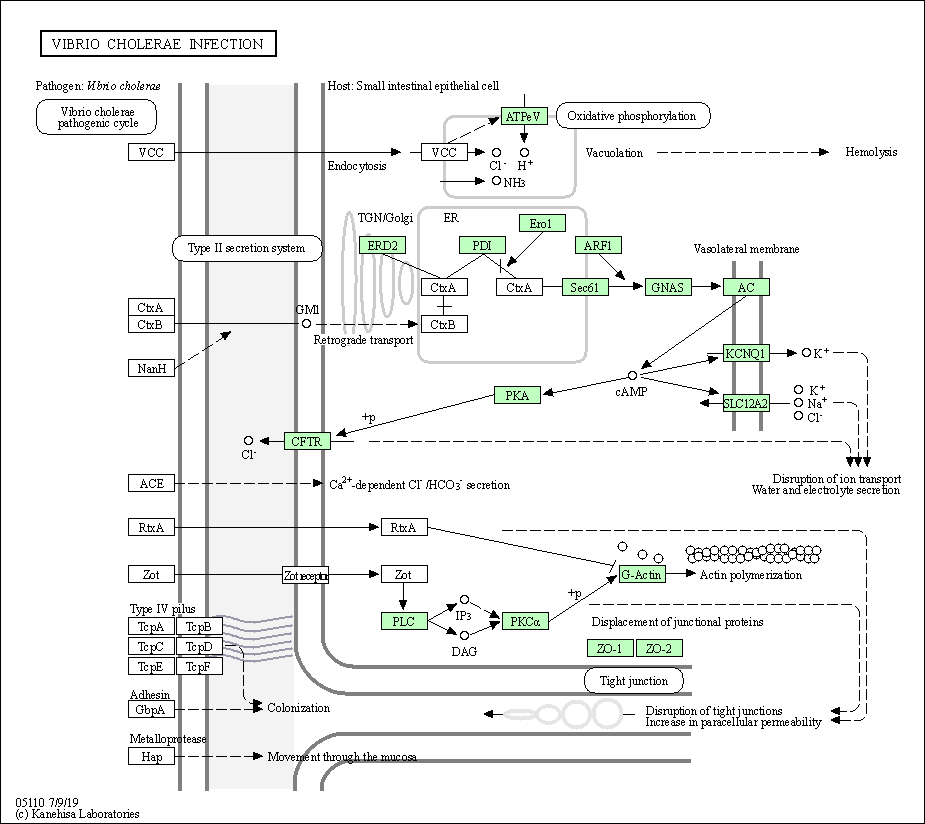
|
|||||||
| Amoebiasis | hsa05146 |
Pathway Map 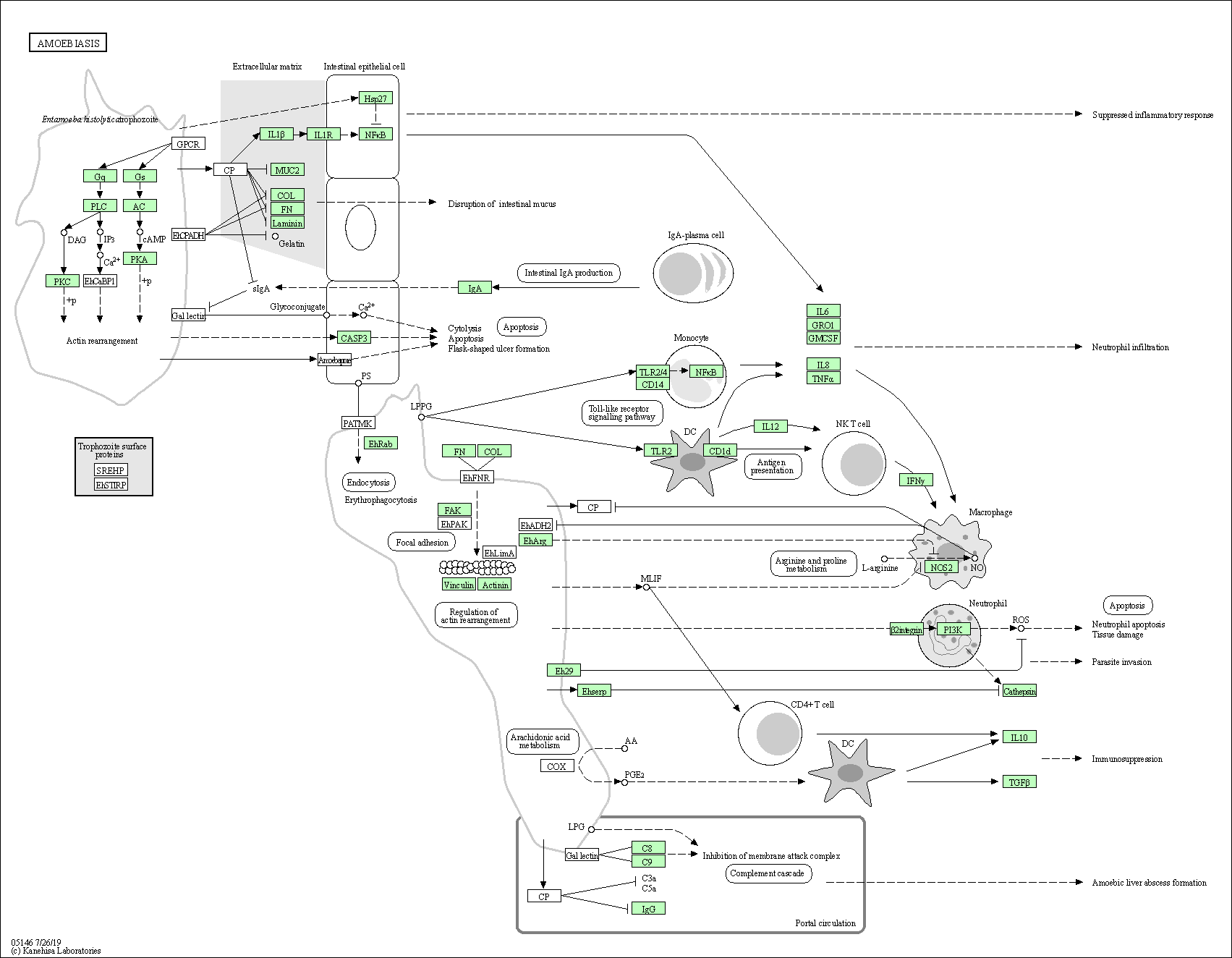
|
|||||||
| Human cytomegalovirus infection | hsa05163 |
Pathway Map 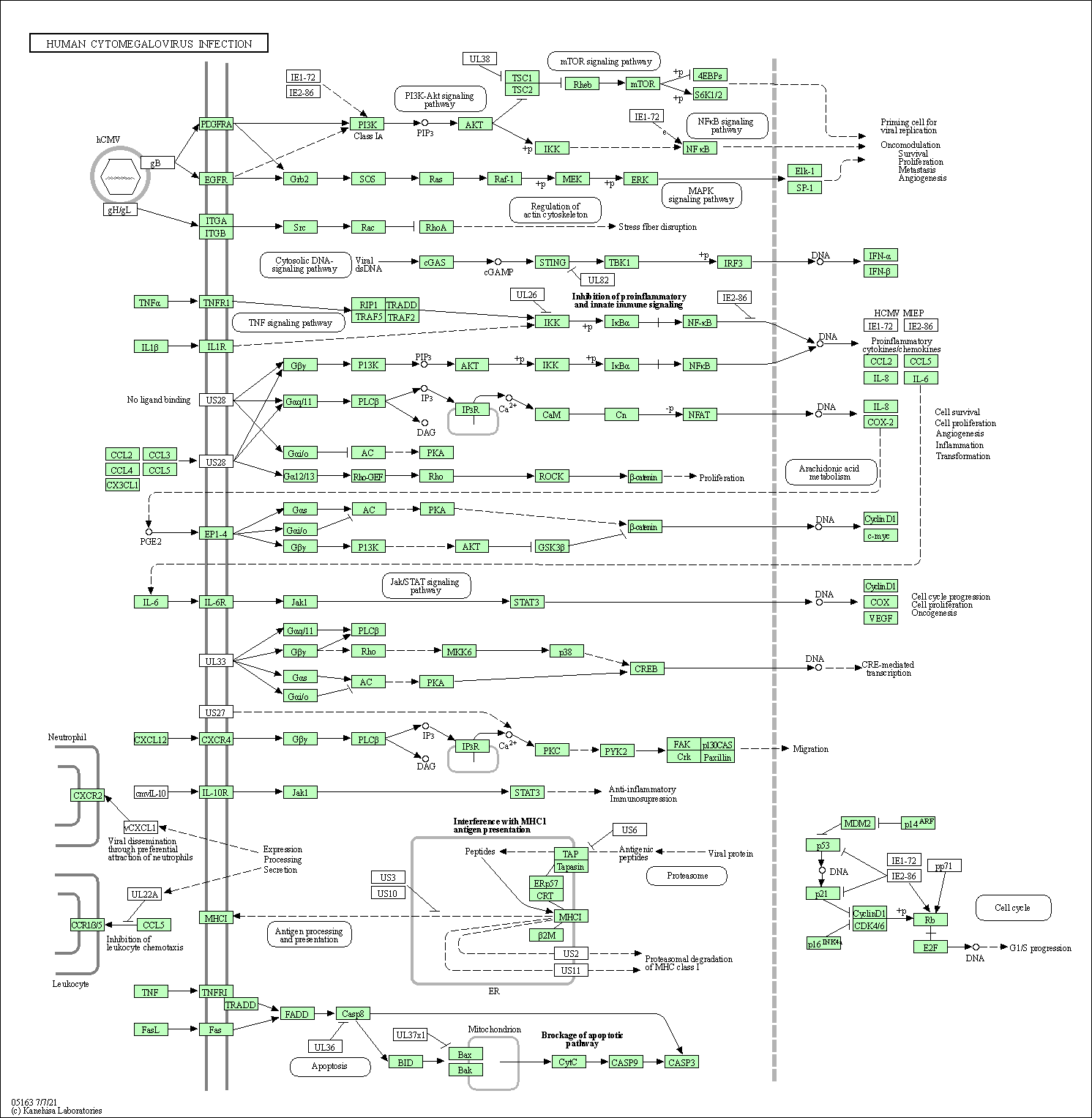
|
|||||||
| Human papillomavirus infection | hsa05165 |
Pathway Map 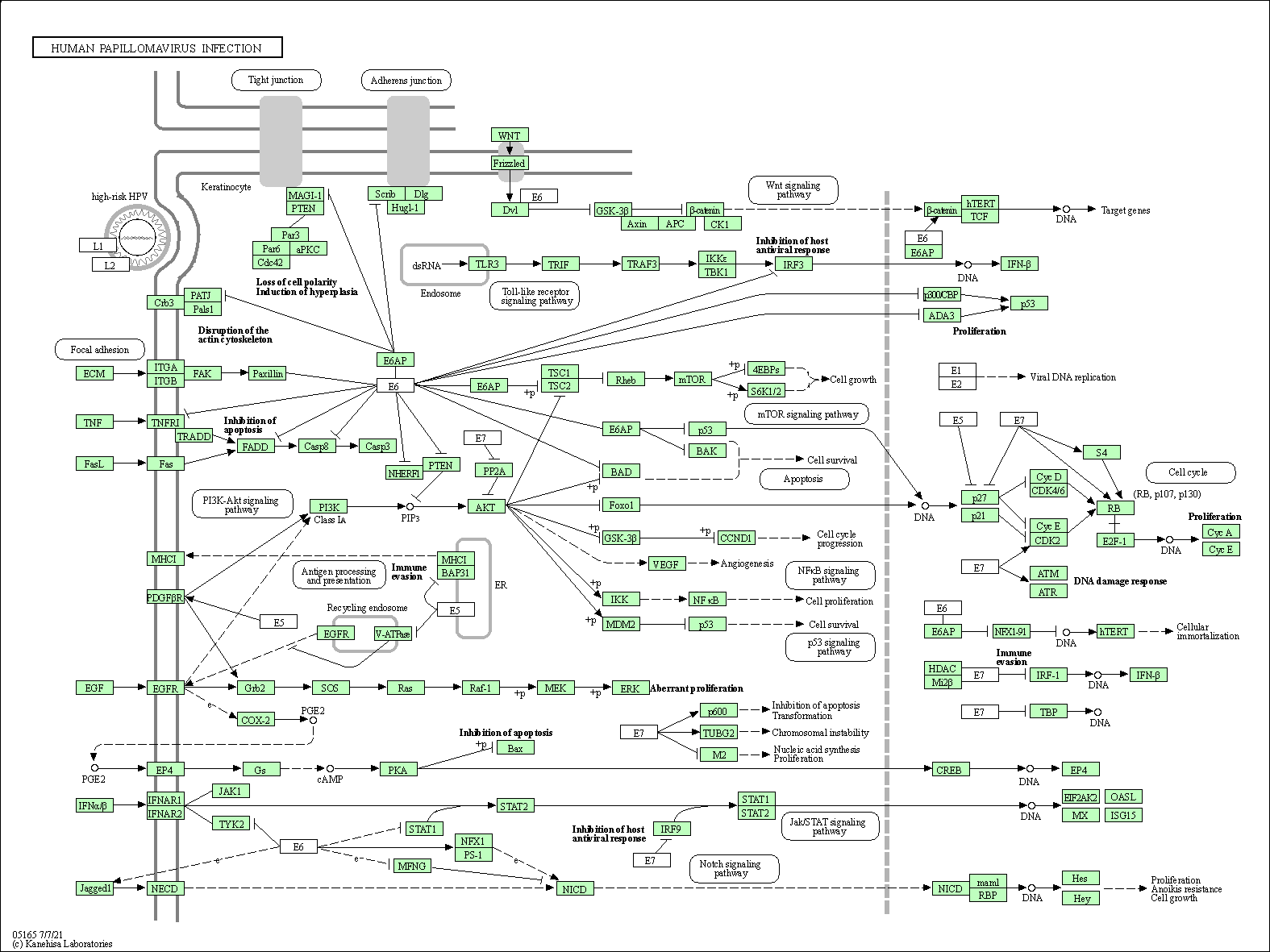
|
|||||||
| Human T-cell leukemia virus 1 infection | hsa05166 |
Pathway Map 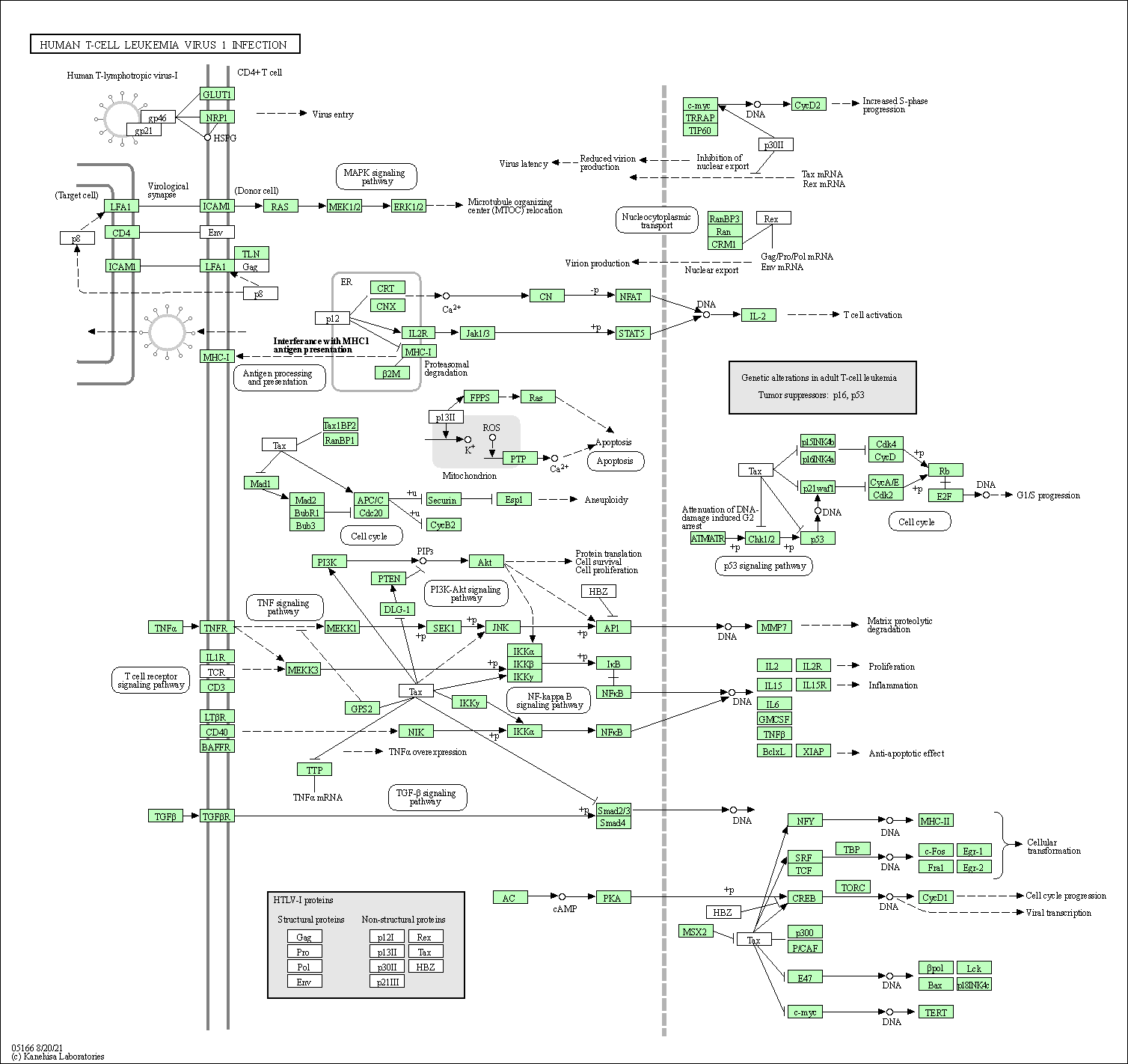
|
|||||||
| Endocrine resistance | hsa01522 |
Pathway Map 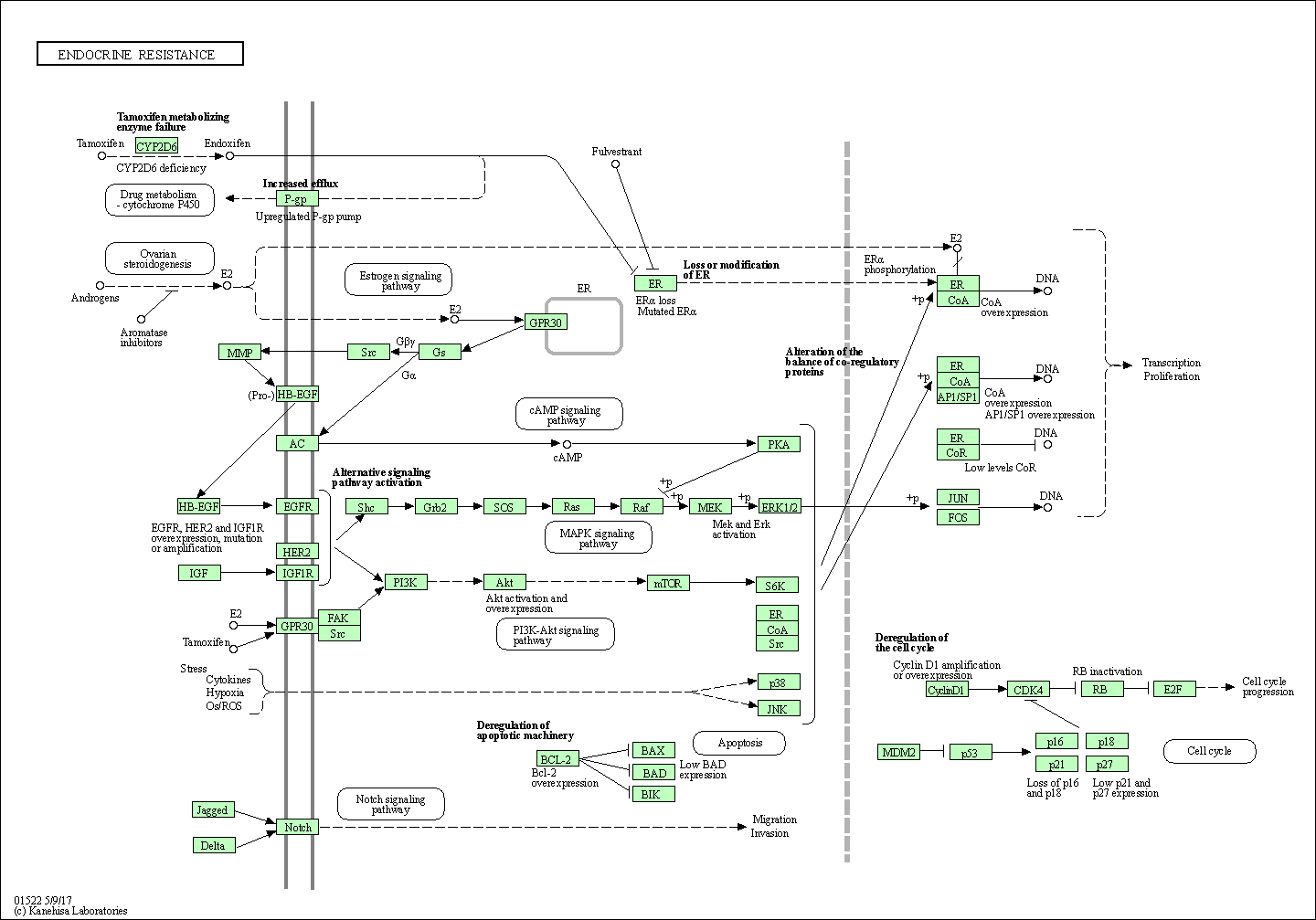
|
|||||||
| Longevity regulating pathway | hsa04211 |
Pathway Map 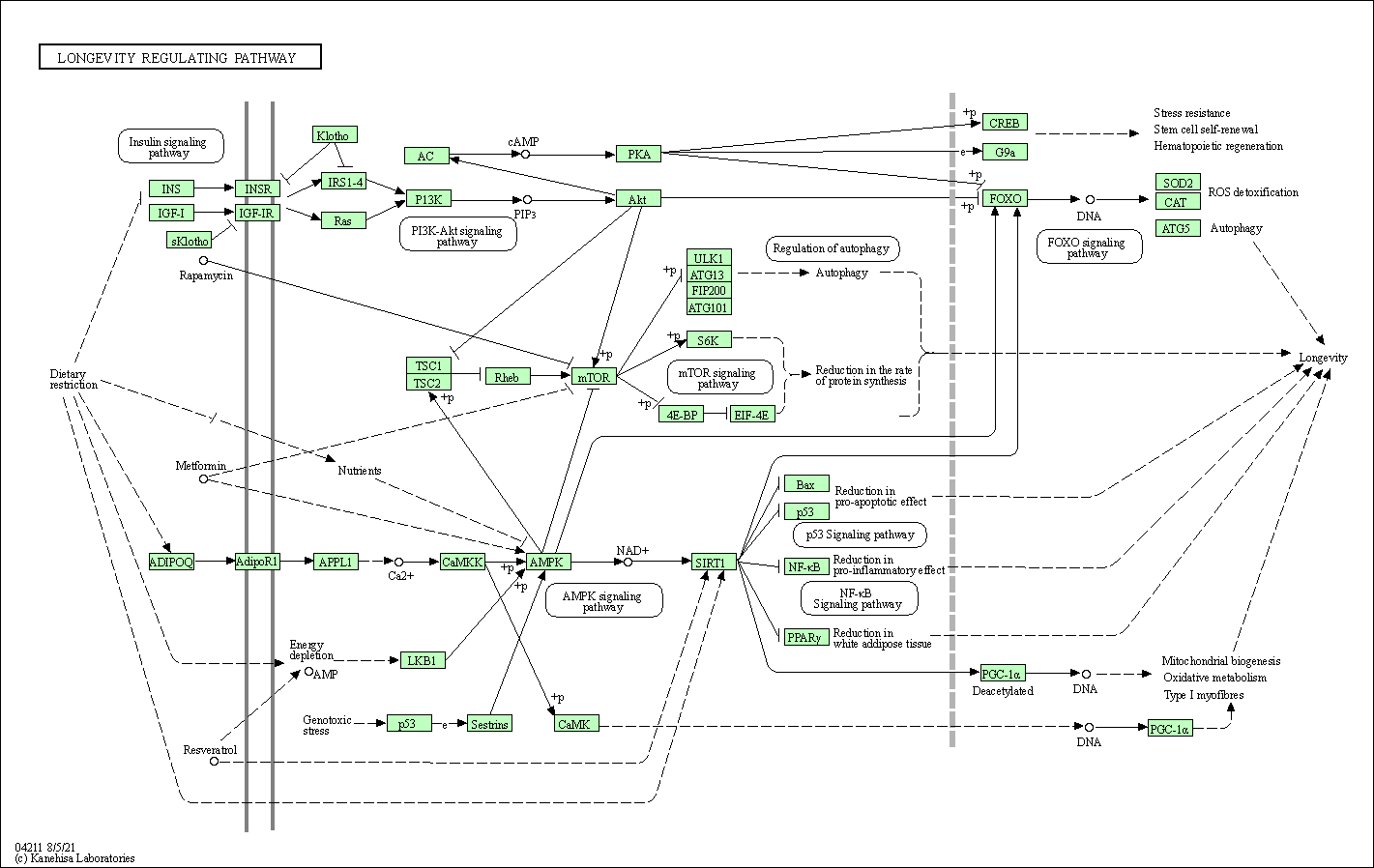
|
|||||||
| Longevity regulating pathway - multiple species | hsa04213 |
Pathway Map 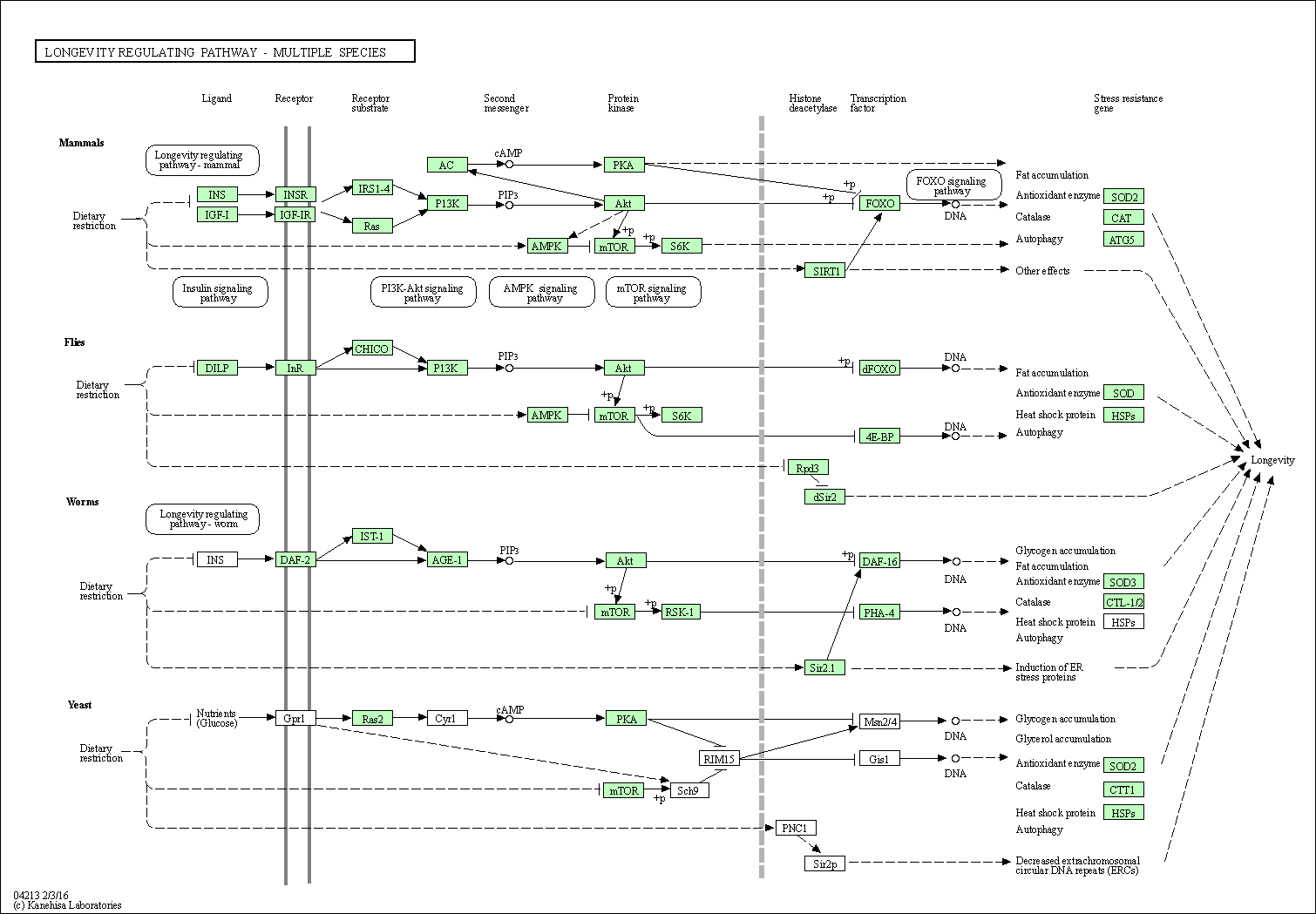
|
|||||||
| Pathways in cancer | hsa05200 |
Pathway Map 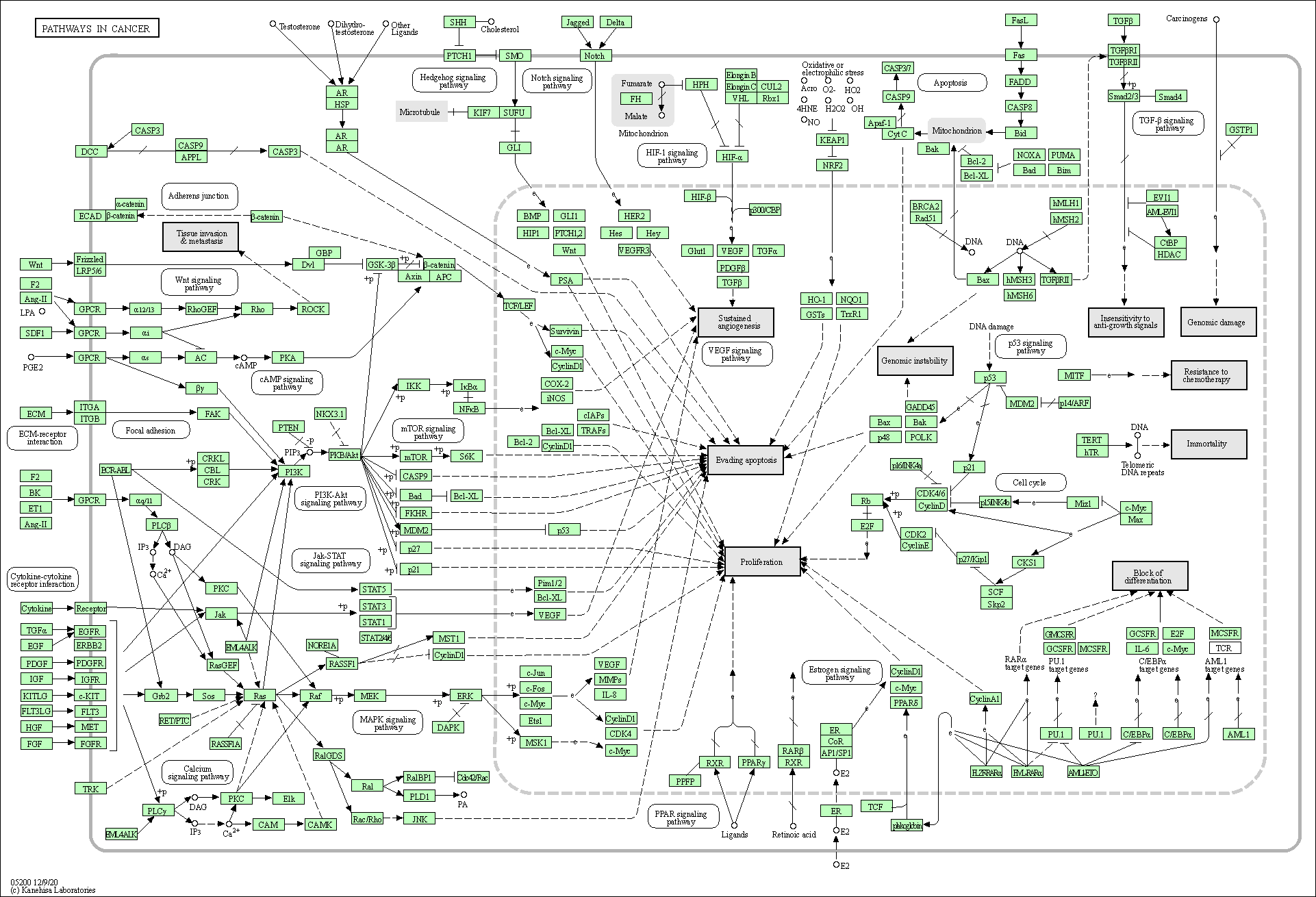
|
|||||||
| Viral carcinogenesis | hsa05203 |
Pathway Map 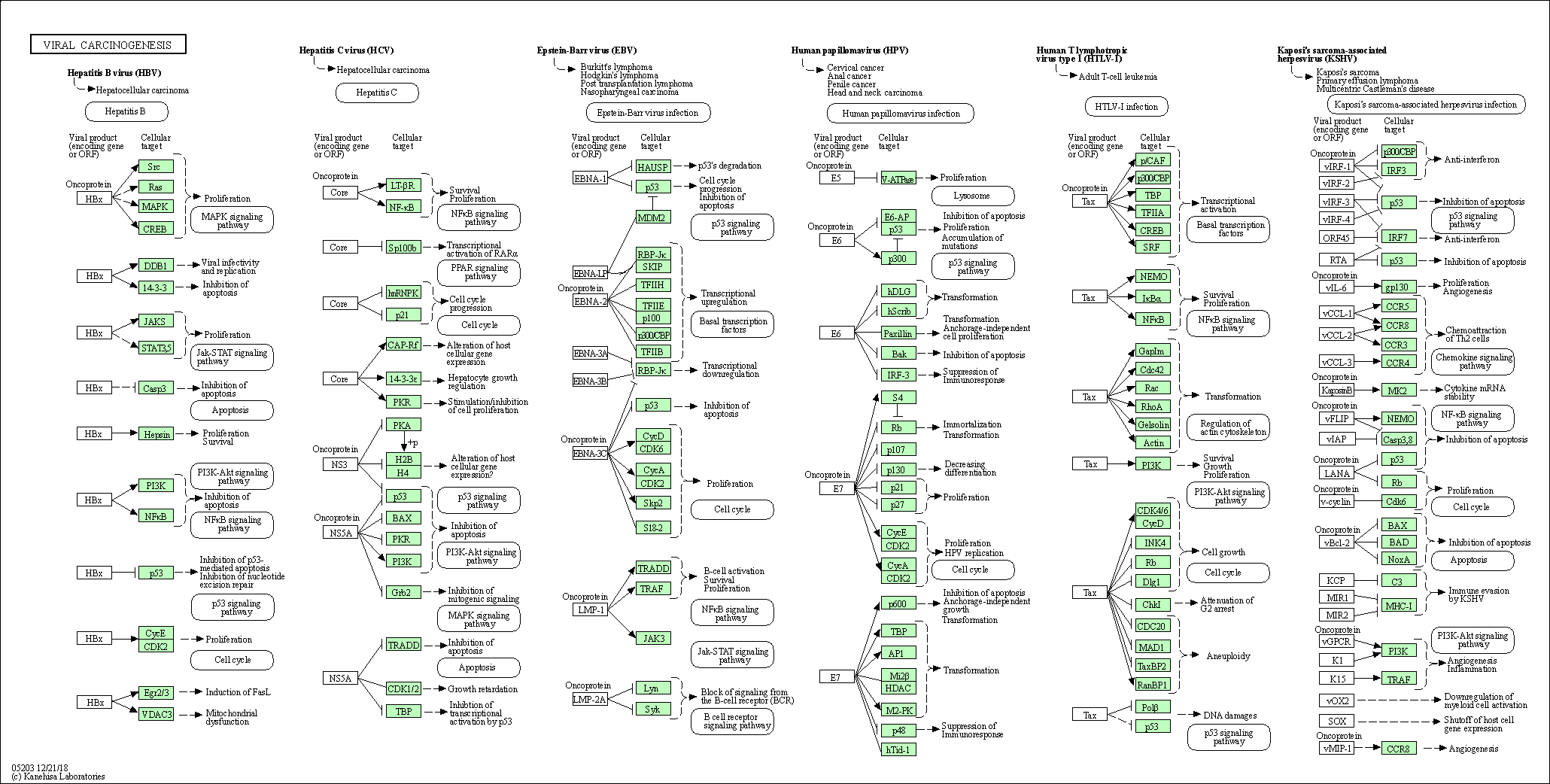
|
|||||||
| Proteoglycans in cancer | hsa05205 |
Pathway Map 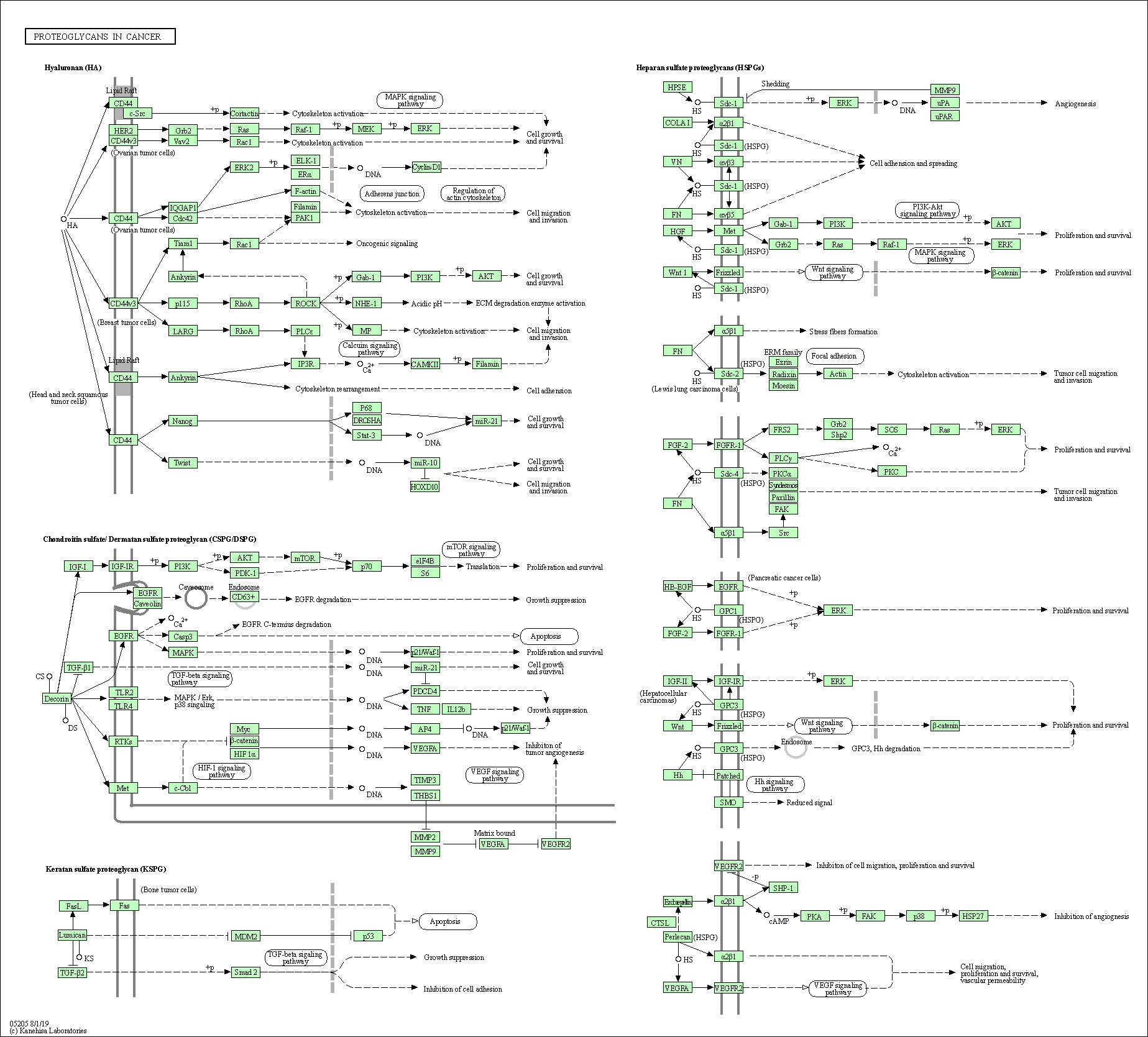
|
|||||||
| Chemical carcinogenesis - receptor activation | hsa05207 |
Pathway Map 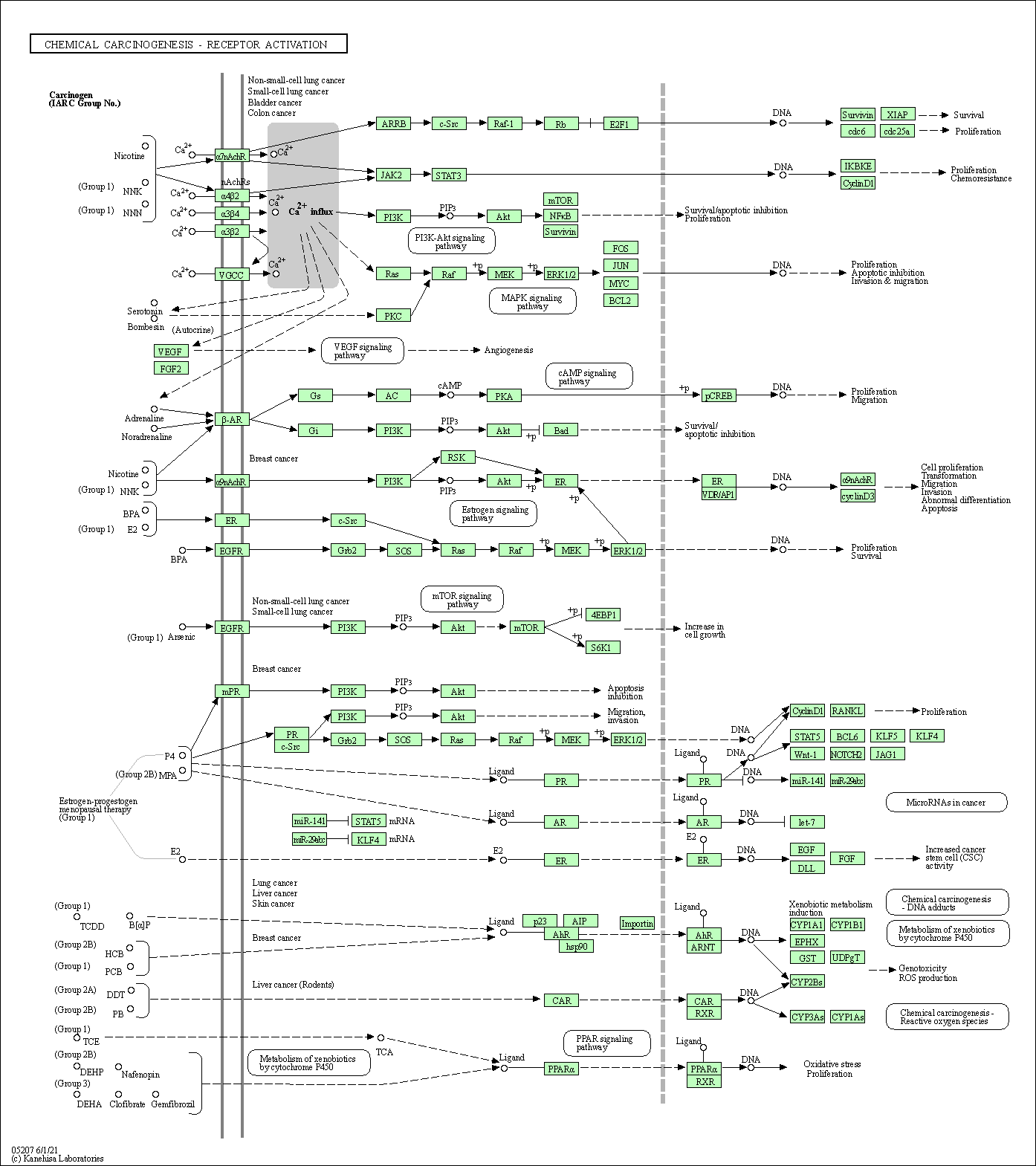
|
|||||||
| Dilated cardiomyopathy | hsa05414 |
Pathway Map 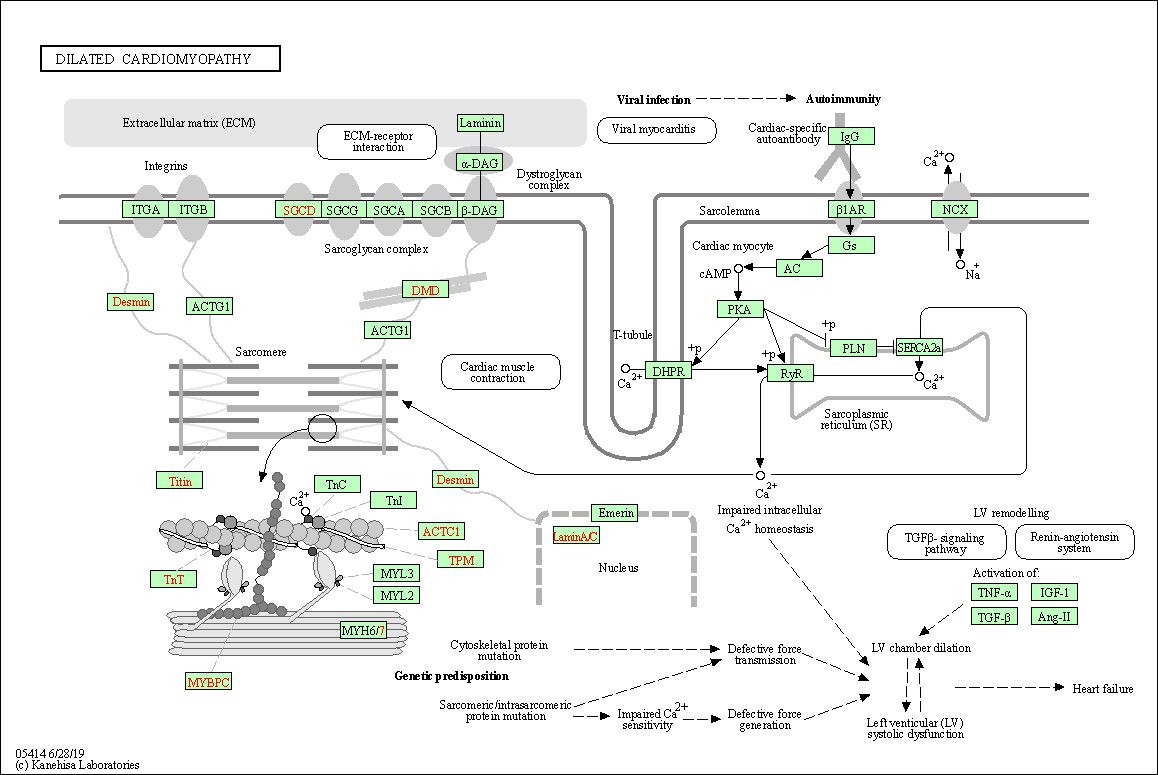
|
|||||||
| Oocyte meiosis | hsa04114 |
Pathway Map 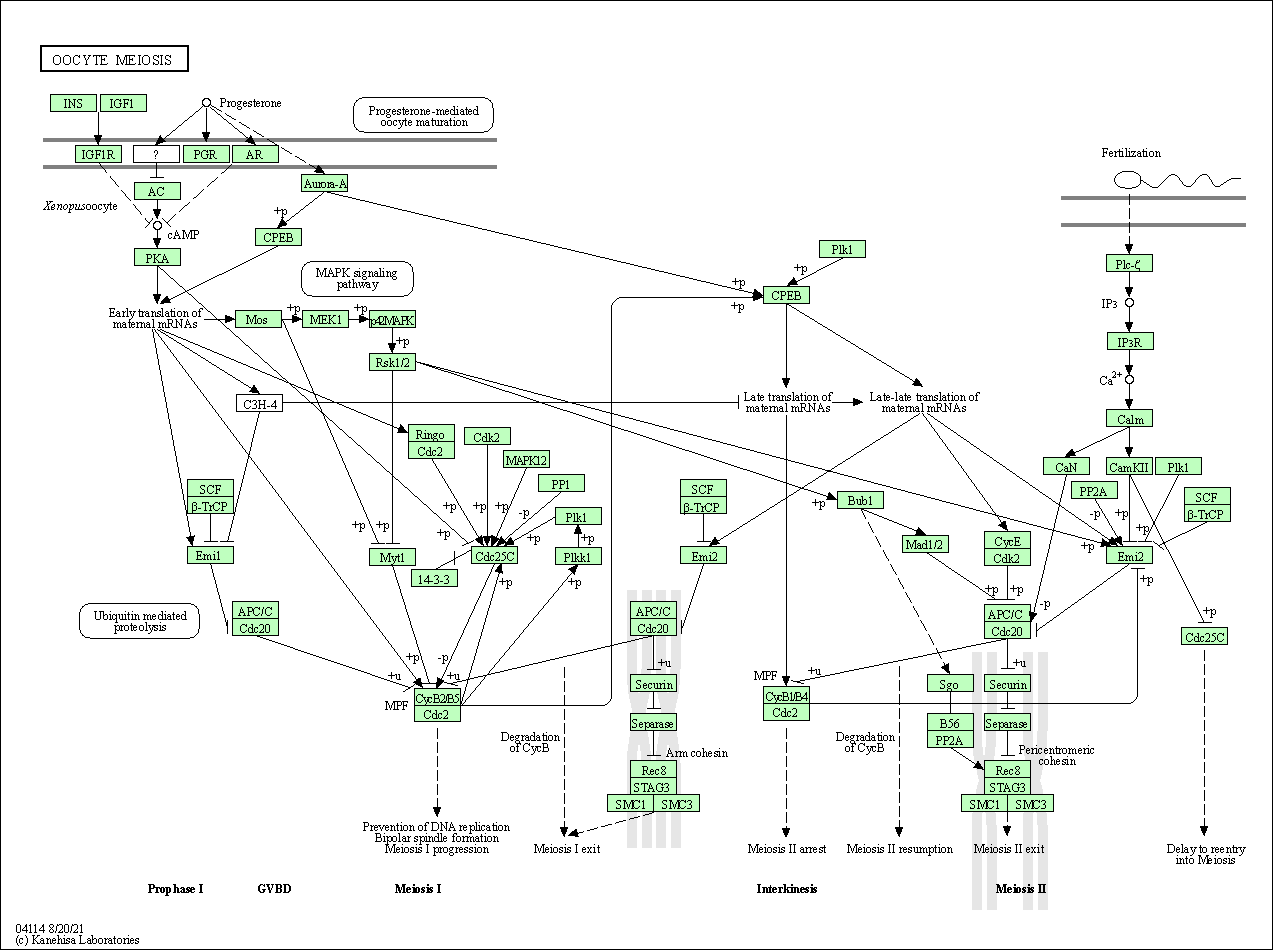
|
|||||||
| Tight junction | hsa04530 |
Pathway Map 
|
|||||||
| Gap junction | hsa04540 |
Pathway Map 
|
|||||||
| Adrenergic signaling in cardiomyocytes | hsa04261 |
Pathway Map 
|
|||||||
| Vascular smooth muscle contraction | hsa04270 |
Pathway Map 
|
|||||||
| Salivary secretion | hsa04970 |
Pathway Map 
|
|||||||
| Gastric acid secretion | hsa04971 |
Pathway Map 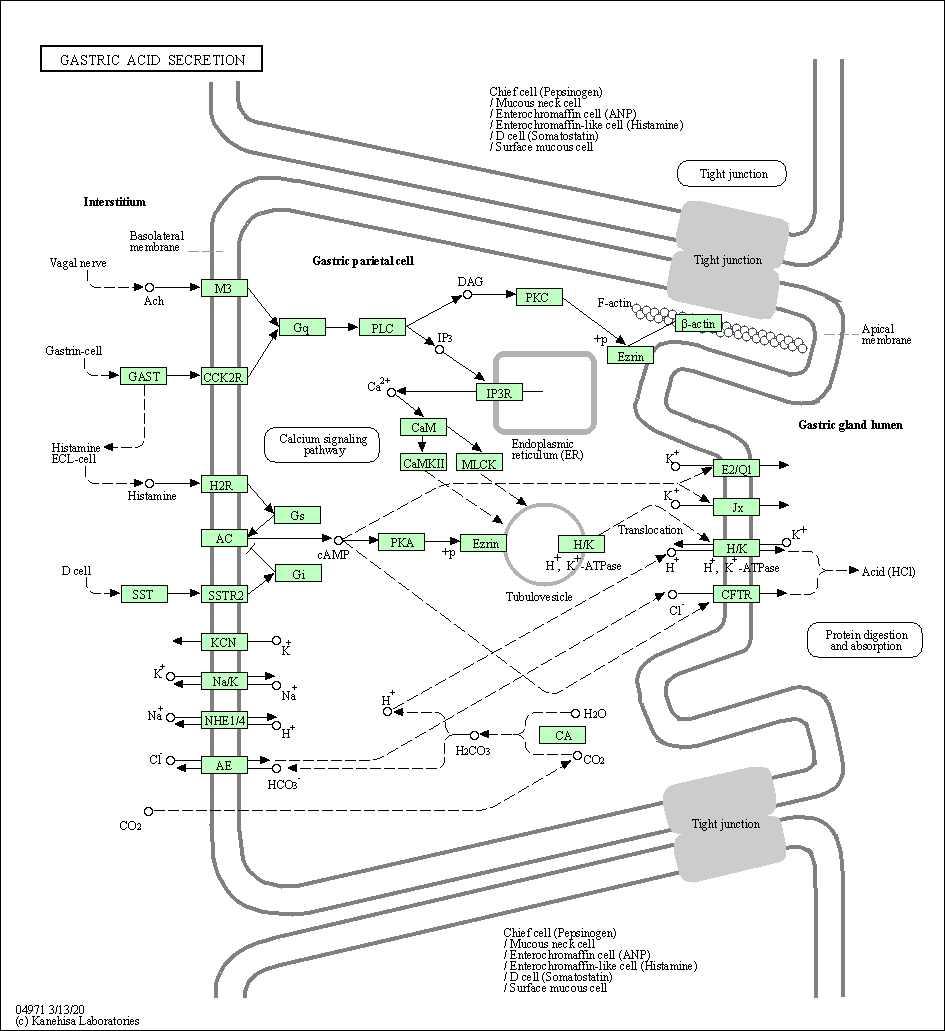
|
|||||||
| Bile secretion | hsa04976 |
Pathway Map 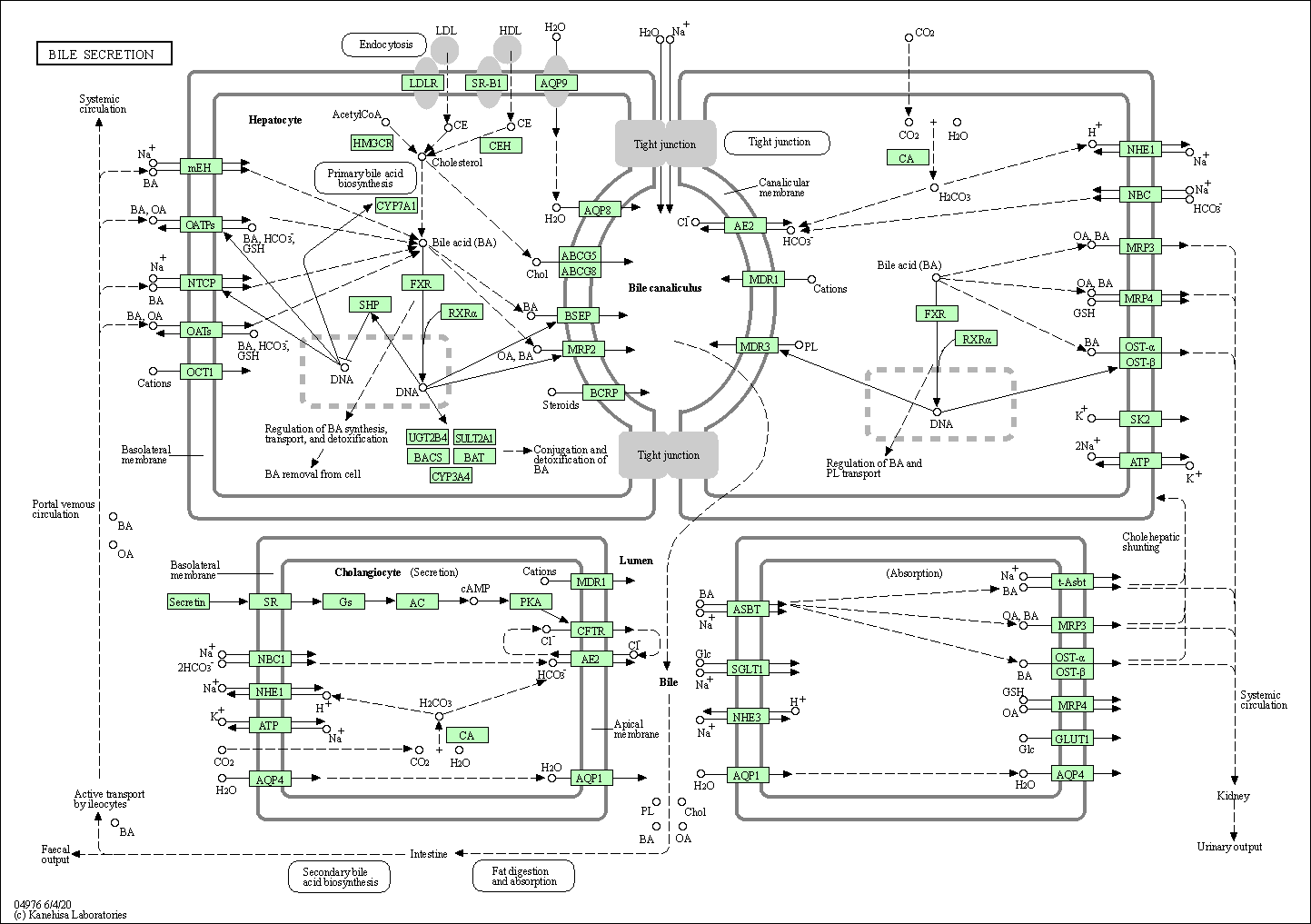
|
|||||||
| Cushing syndrome | hsa04934 |
Pathway Map 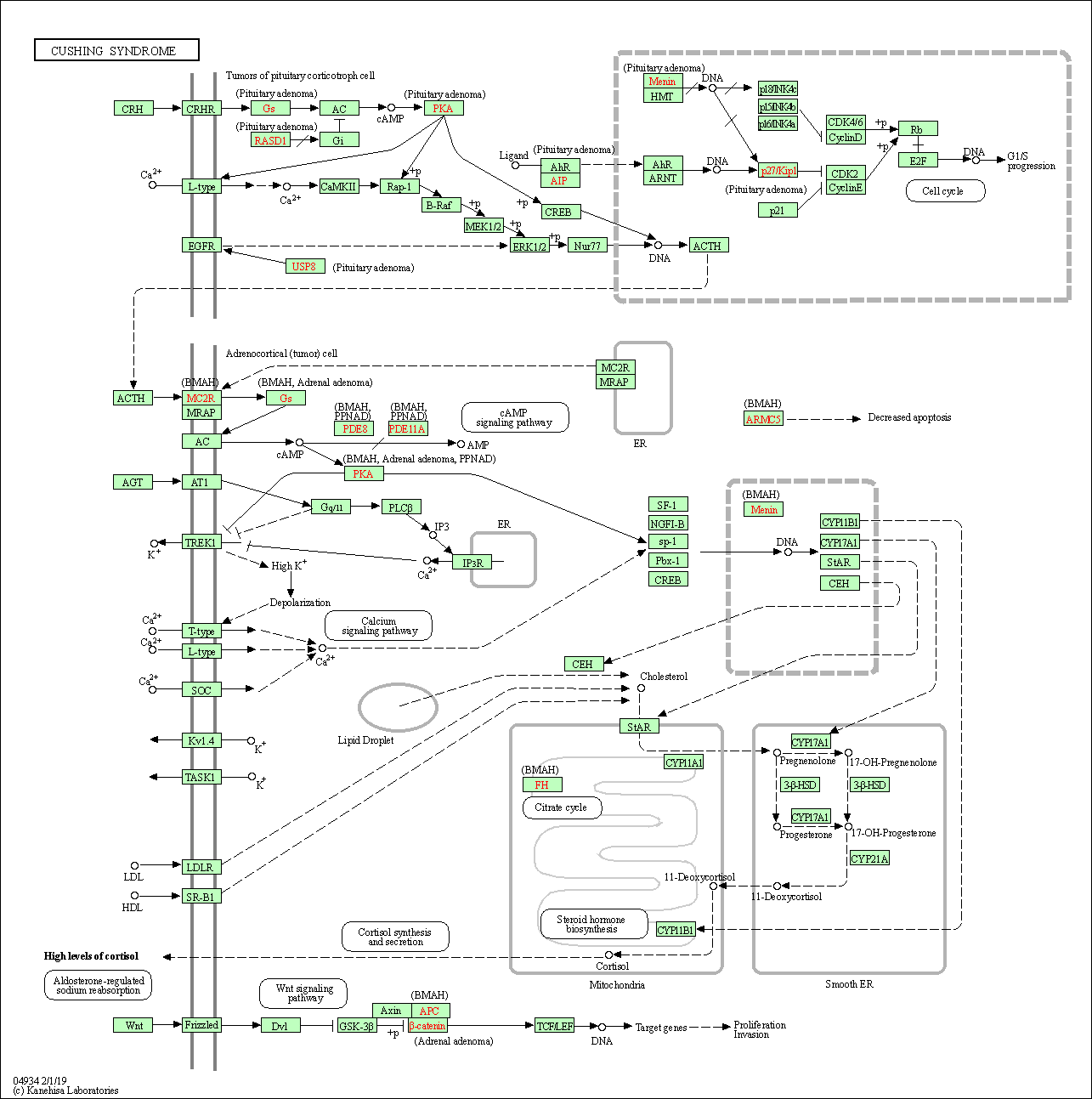
|
|||||||
| Insulin signaling pathway | hsa04910 |
Pathway Map 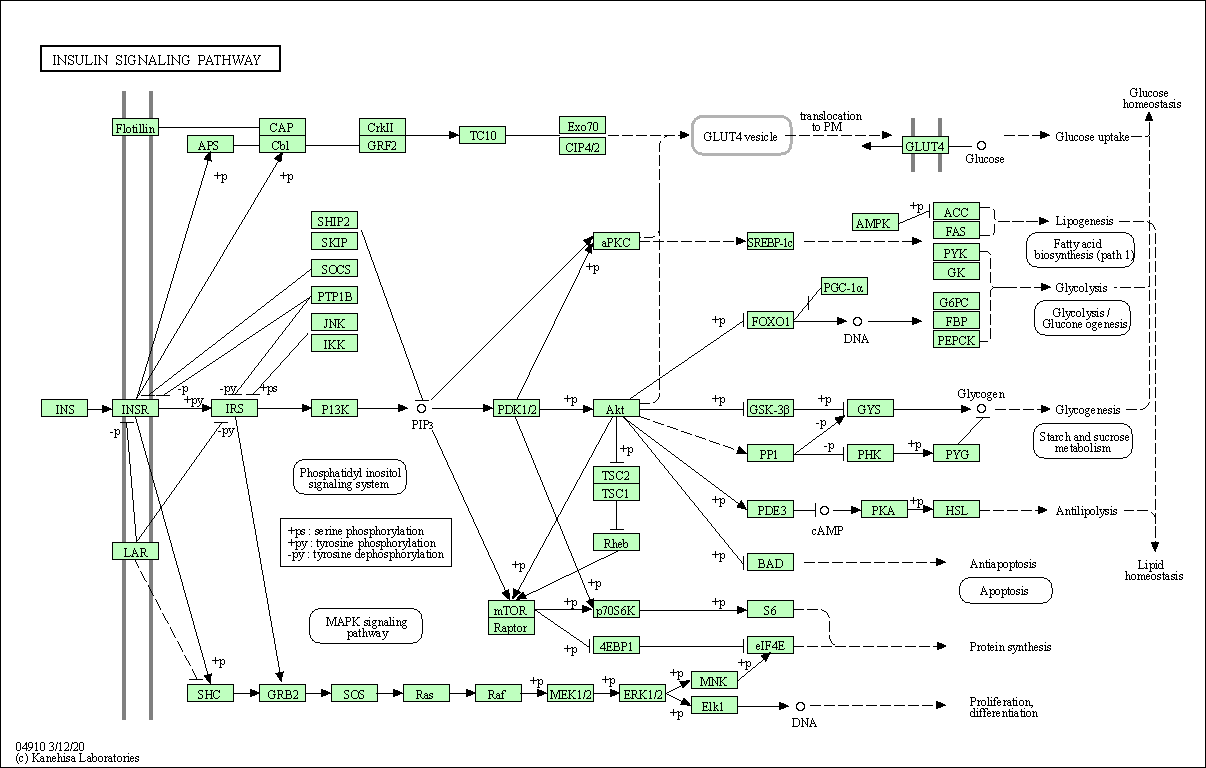
|
|||||||
| Insulin secretion | hsa04911 |
Pathway Map 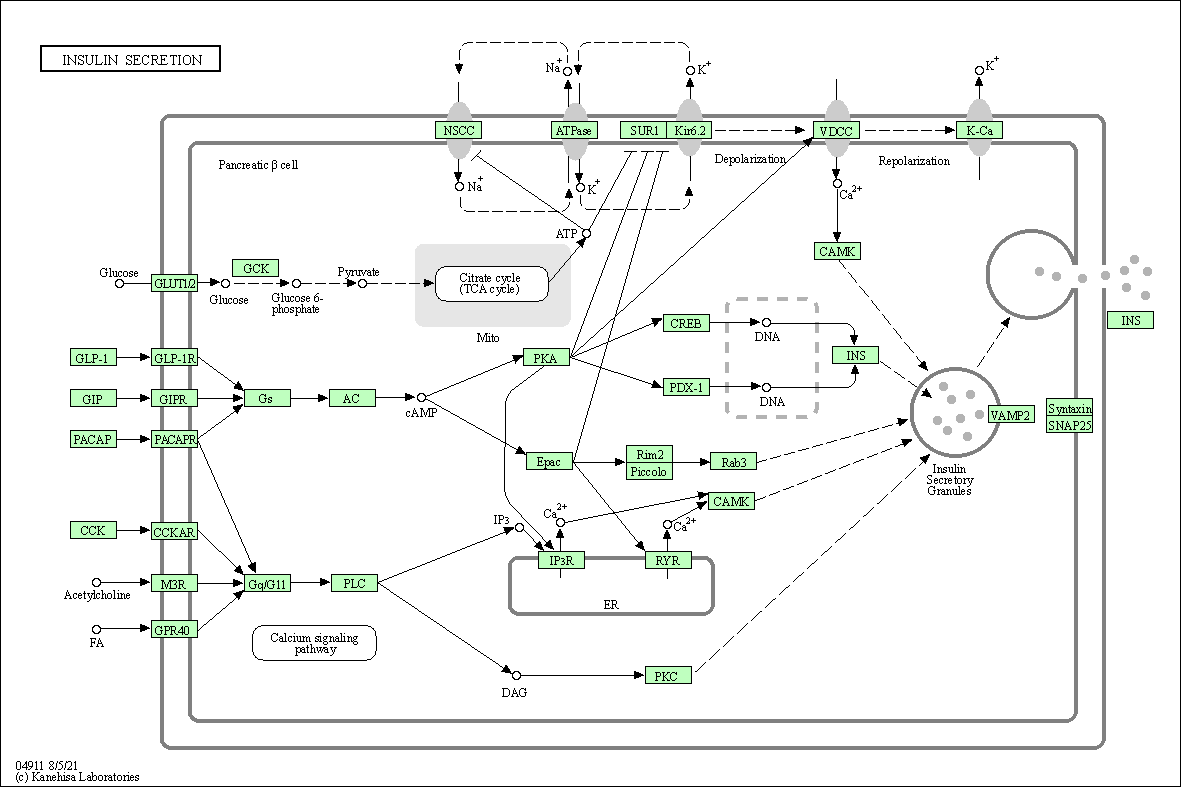
|
|||||||
| GnRH signaling pathway | hsa04912 |
Pathway Map 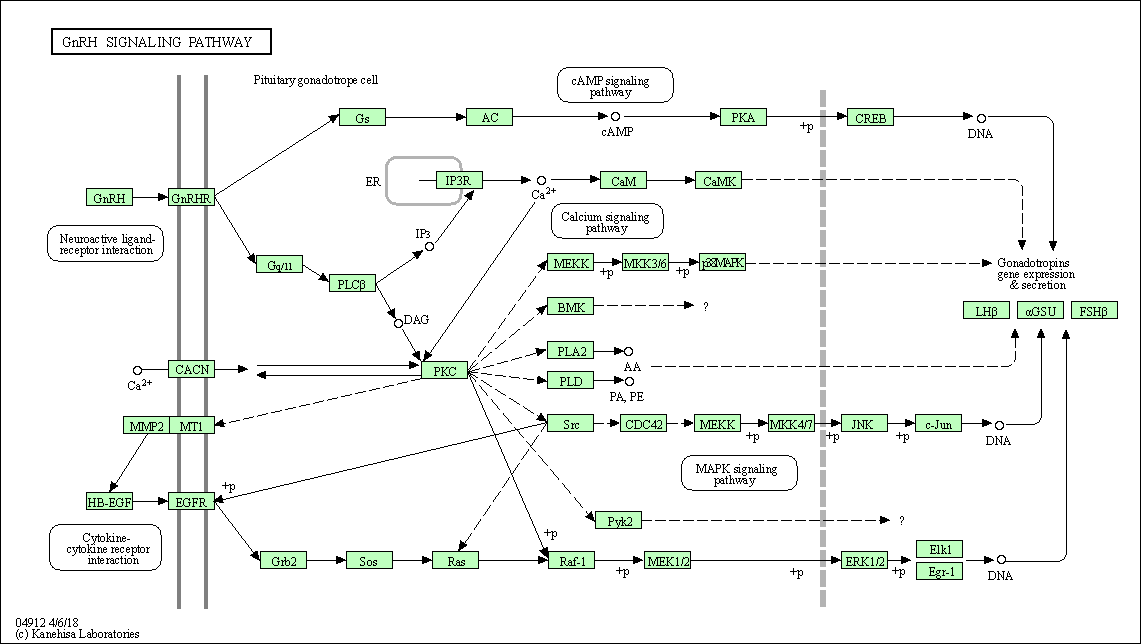
|
|||||||
| Ovarian steroidogenesis | hsa04913 |
Pathway Map 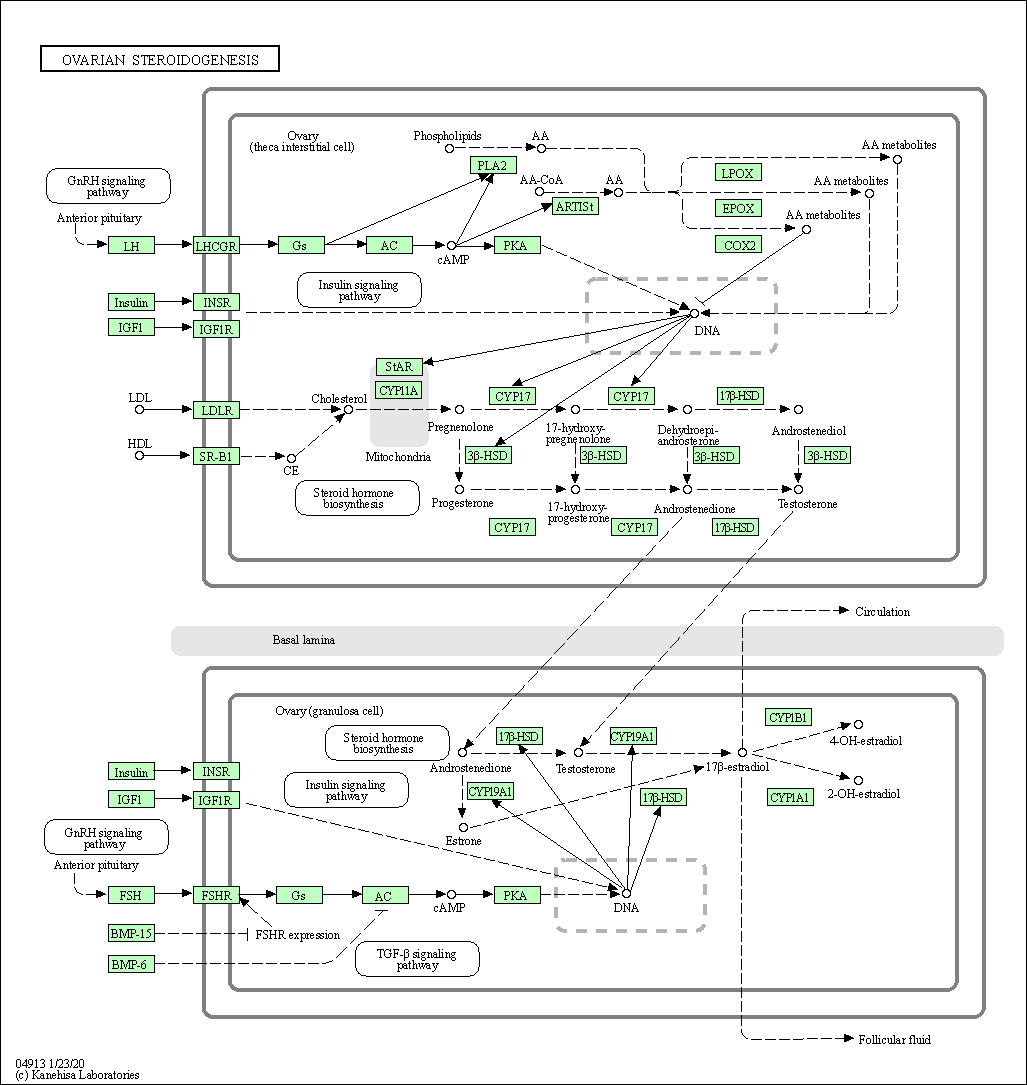
|
|||||||
| Progesterone-mediated oocyte maturation | hsa04914 |
Pathway Map 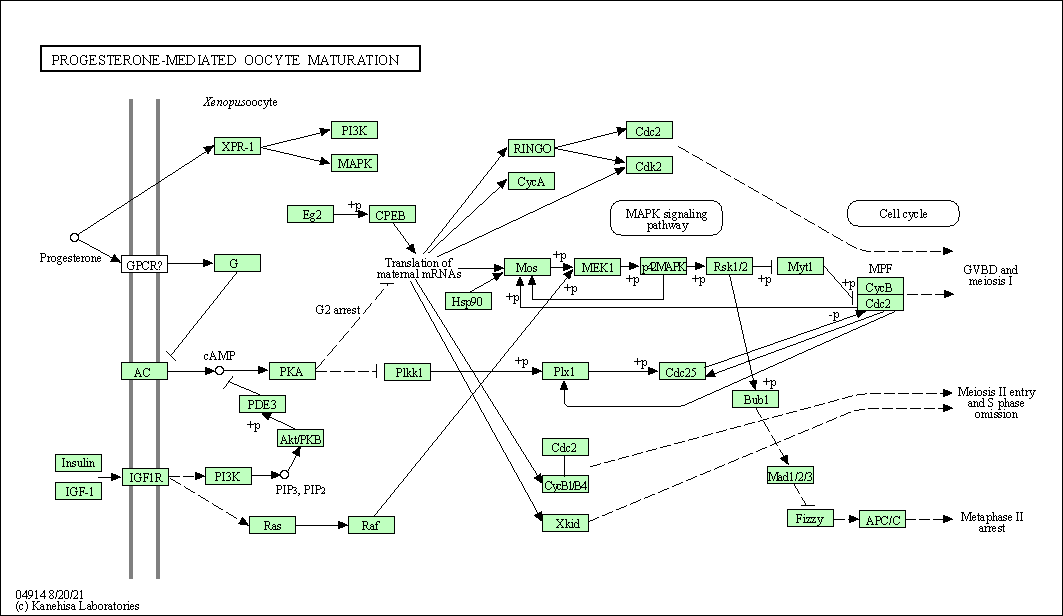
|
|||||||
| Estrogen signaling pathway | hsa04915 |
Pathway Map 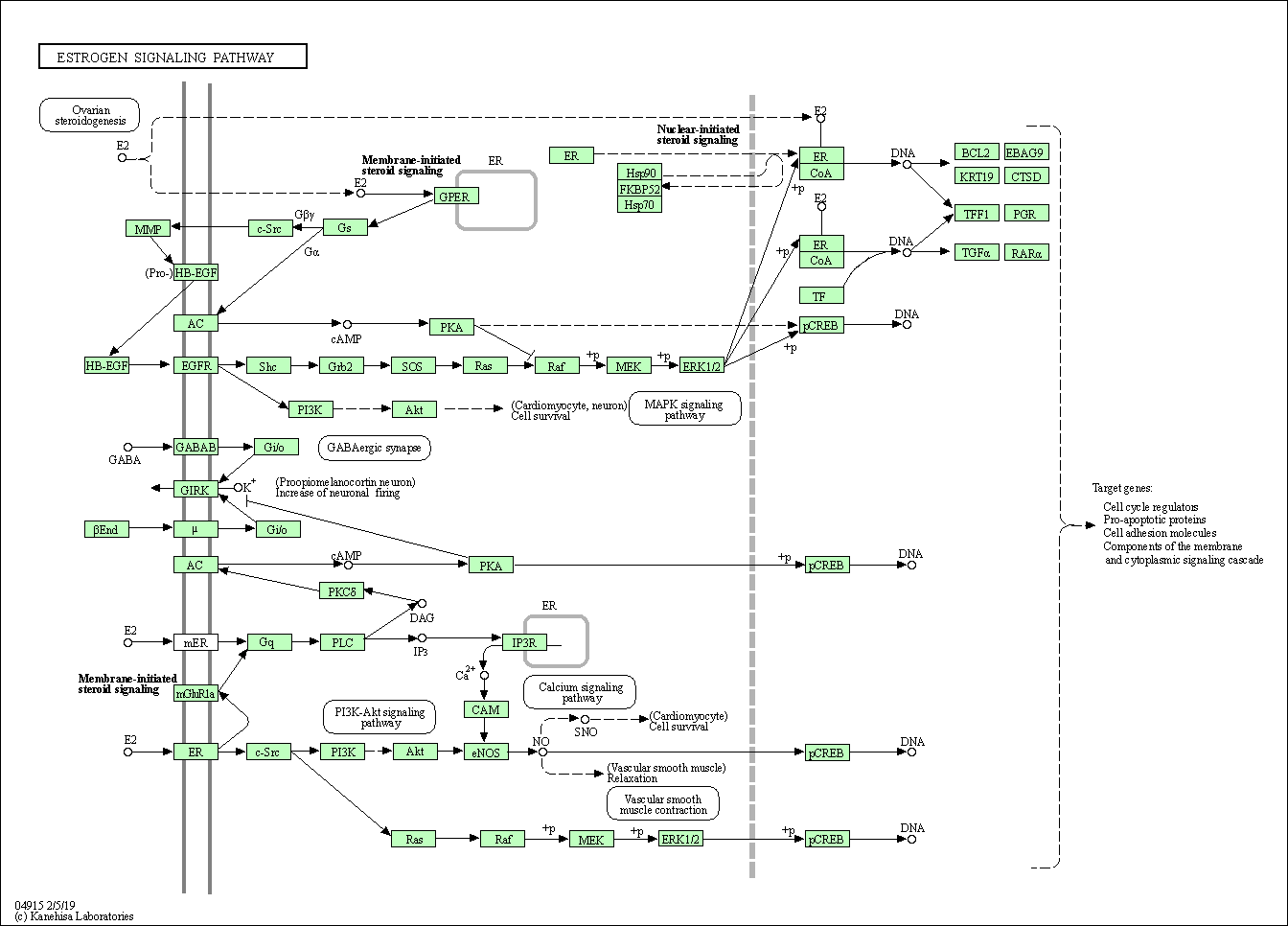
|
|||||||
| Melanogenesis | hsa04916 |
Pathway Map 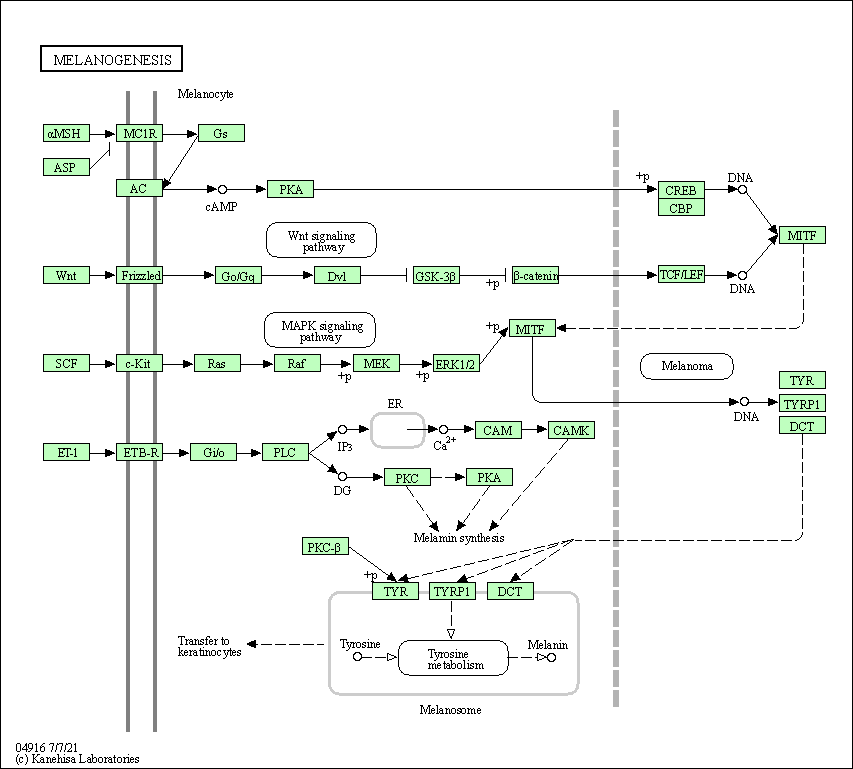
|
|||||||
| Thyroid hormone synthesis | hsa04918 |
Pathway Map 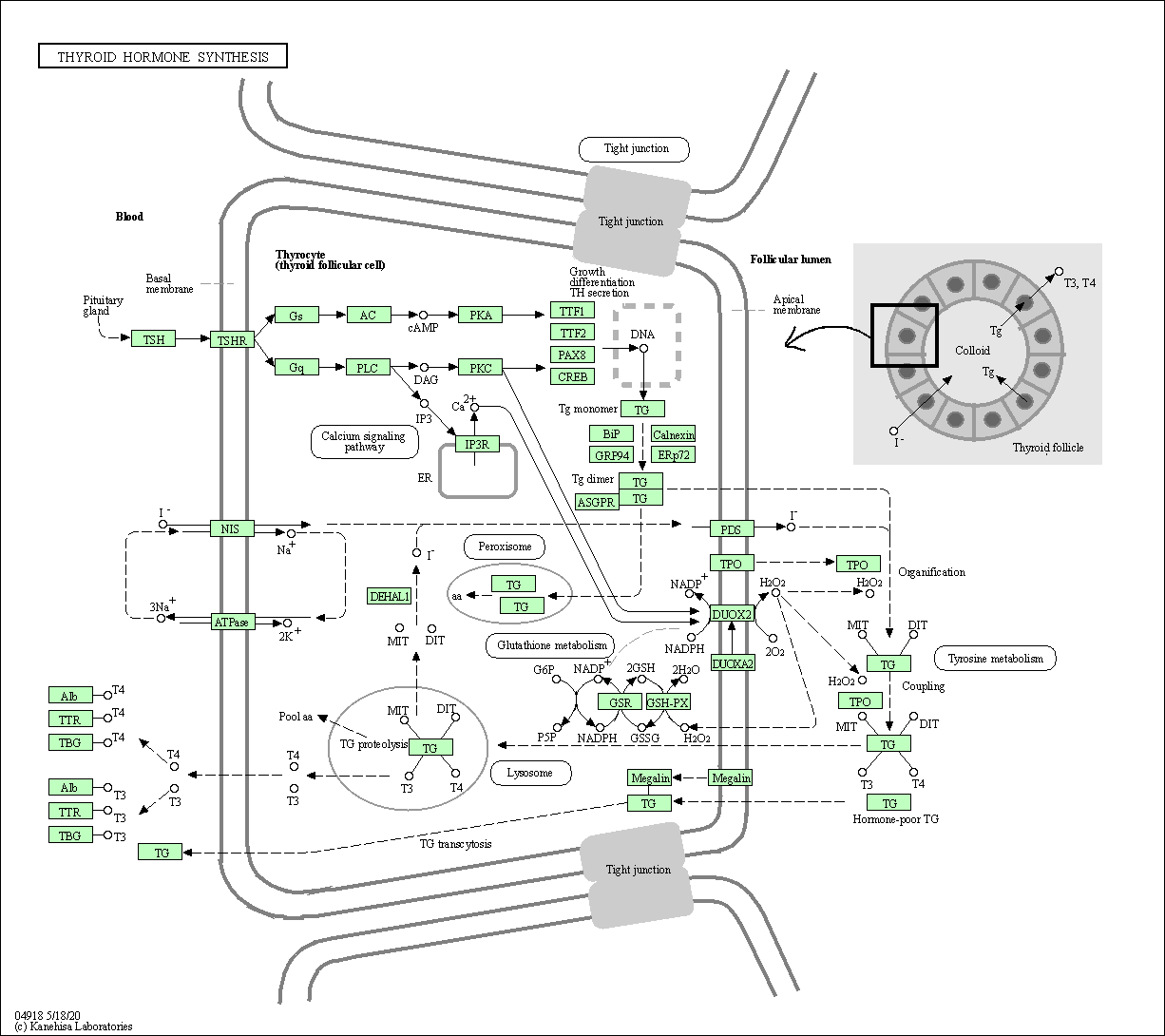
|
|||||||
| Thyroid hormone signaling pathway | hsa04919 |
Pathway Map 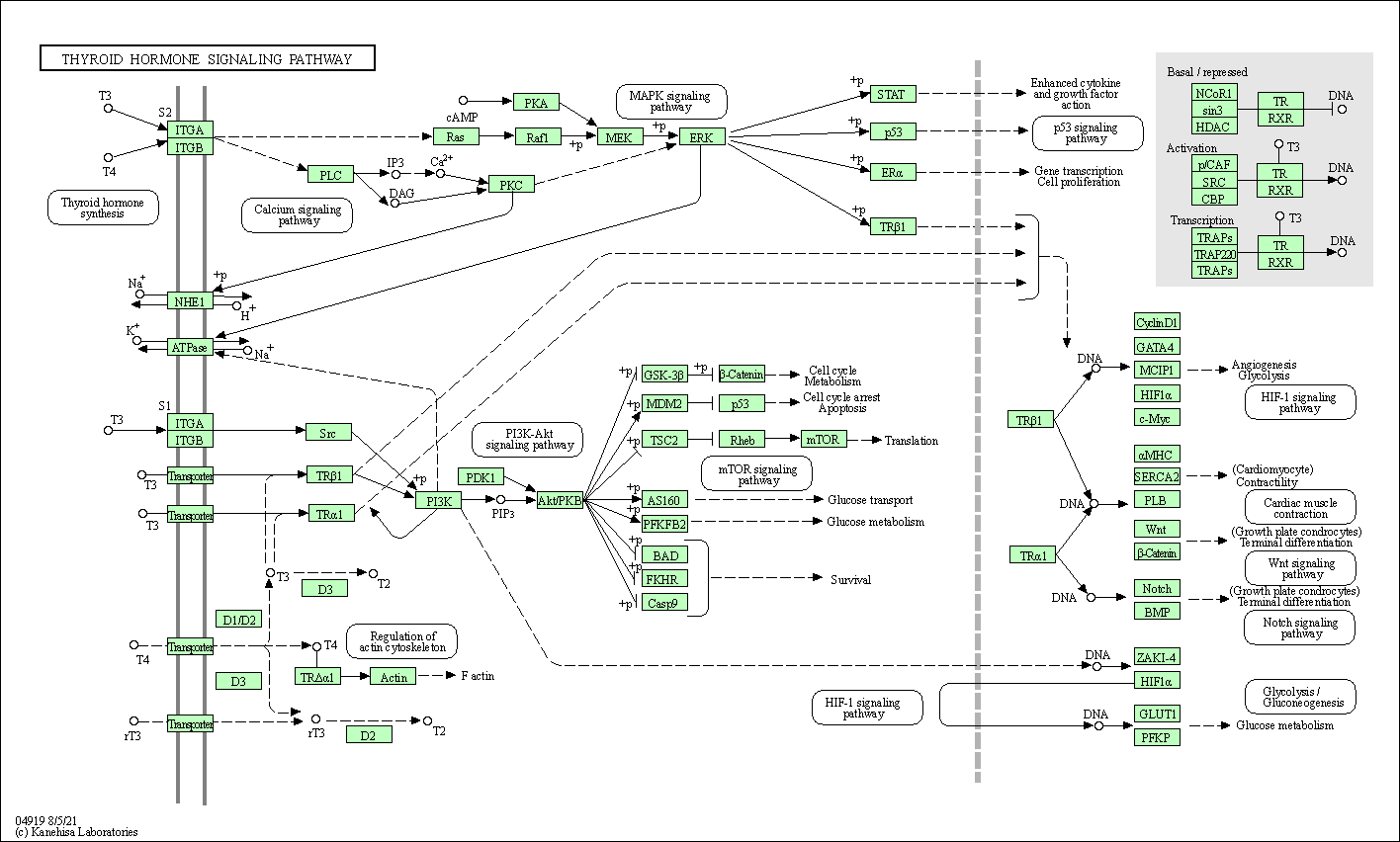
|
|||||||
| Oxytocin signaling pathway | hsa04921 |
Pathway Map 
|
|||||||
| Glucagon signaling pathway | hsa04922 |
Pathway Map 
|
|||||||
| Regulation of lipolysis in adipocytes | hsa04923 |
Pathway Map 
|
|||||||
| Renin secretion | hsa04924 |
Pathway Map 
|
|||||||
| Aldosterone synthesis and secretion | hsa04925 |
Pathway Map 
|
|||||||
| Relaxin signaling pathway | hsa04926 |
Pathway Map 
|
|||||||
| Cortisol synthesis and secretion | hsa04927 |
Pathway Map 
|
|||||||
| Parathyroid hormone synthesis, secretion and action | hsa04928 |
Pathway Map 
|
|||||||
| Growth hormone synthesis, secretion and action | hsa04935 |
Pathway Map 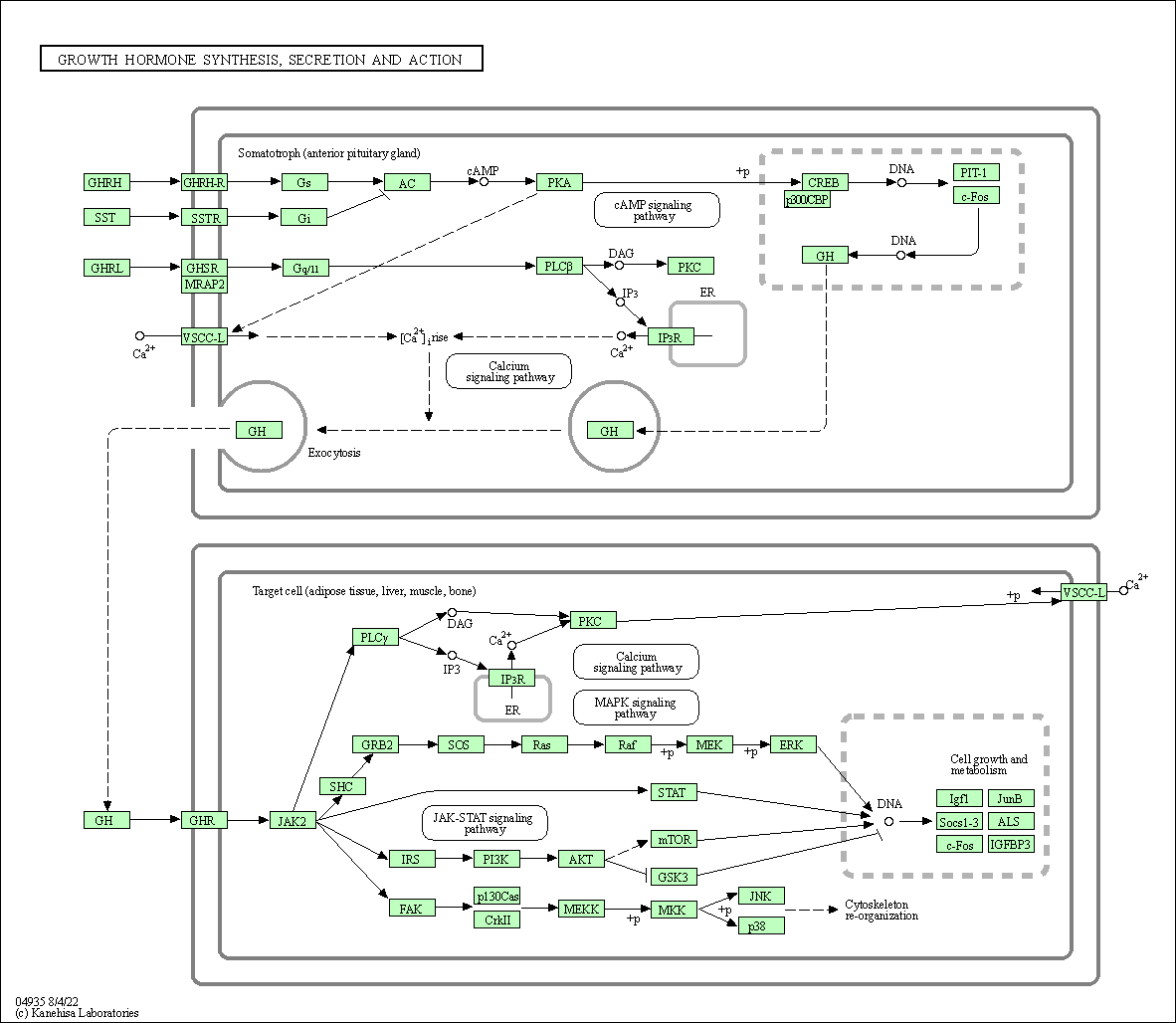
|
|||||||
| Circadian entrainment | hsa04713 |
Pathway Map 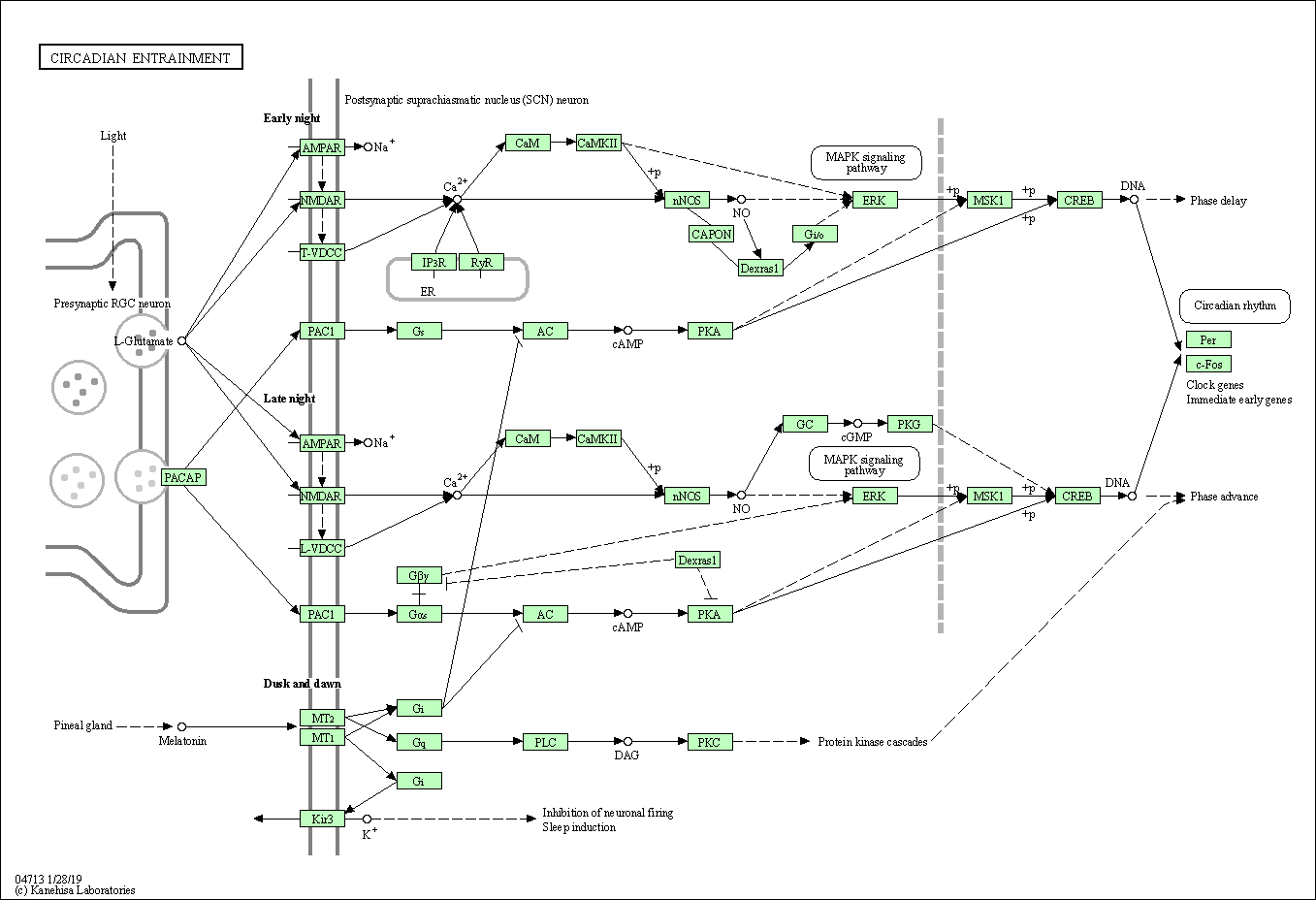
|
|||||||
| Thermogenesis | hsa04714 |
Pathway Map 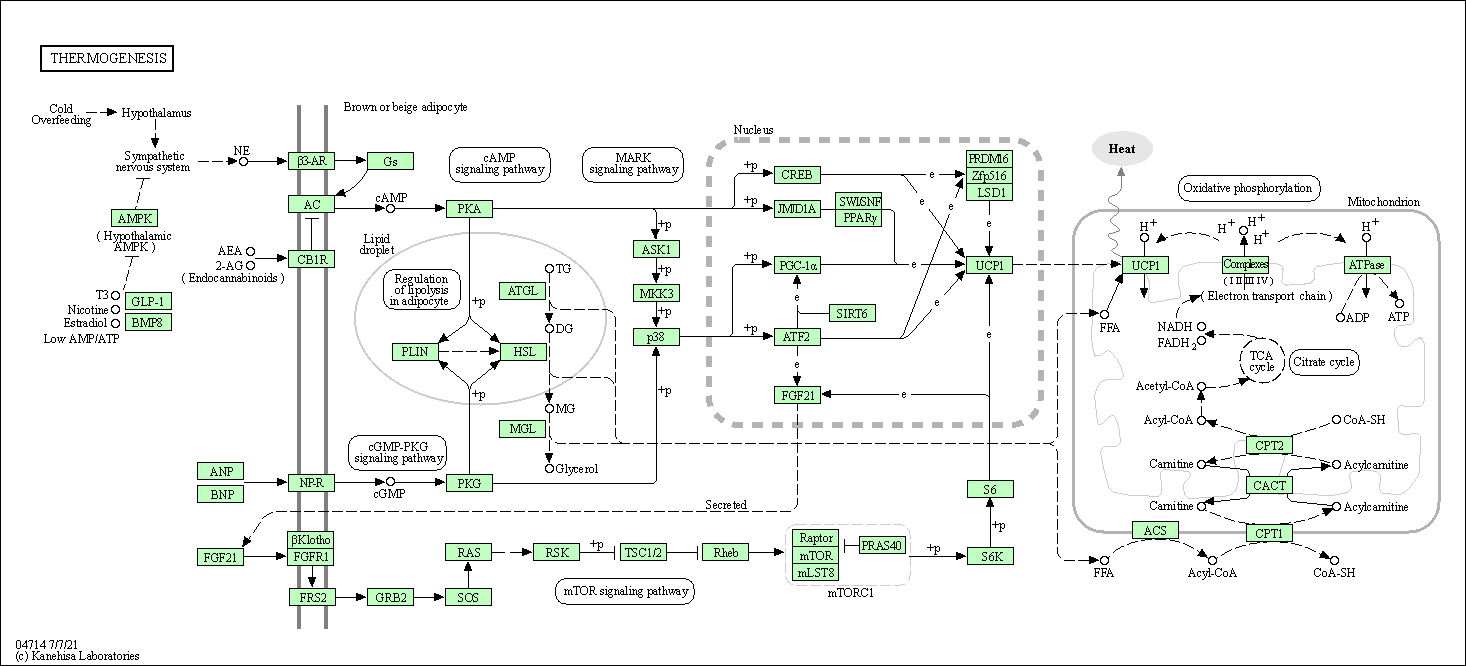
|
|||||||
| Endocrine and other factor-regulated calcium reabsorption | hsa04961 |
Pathway Map 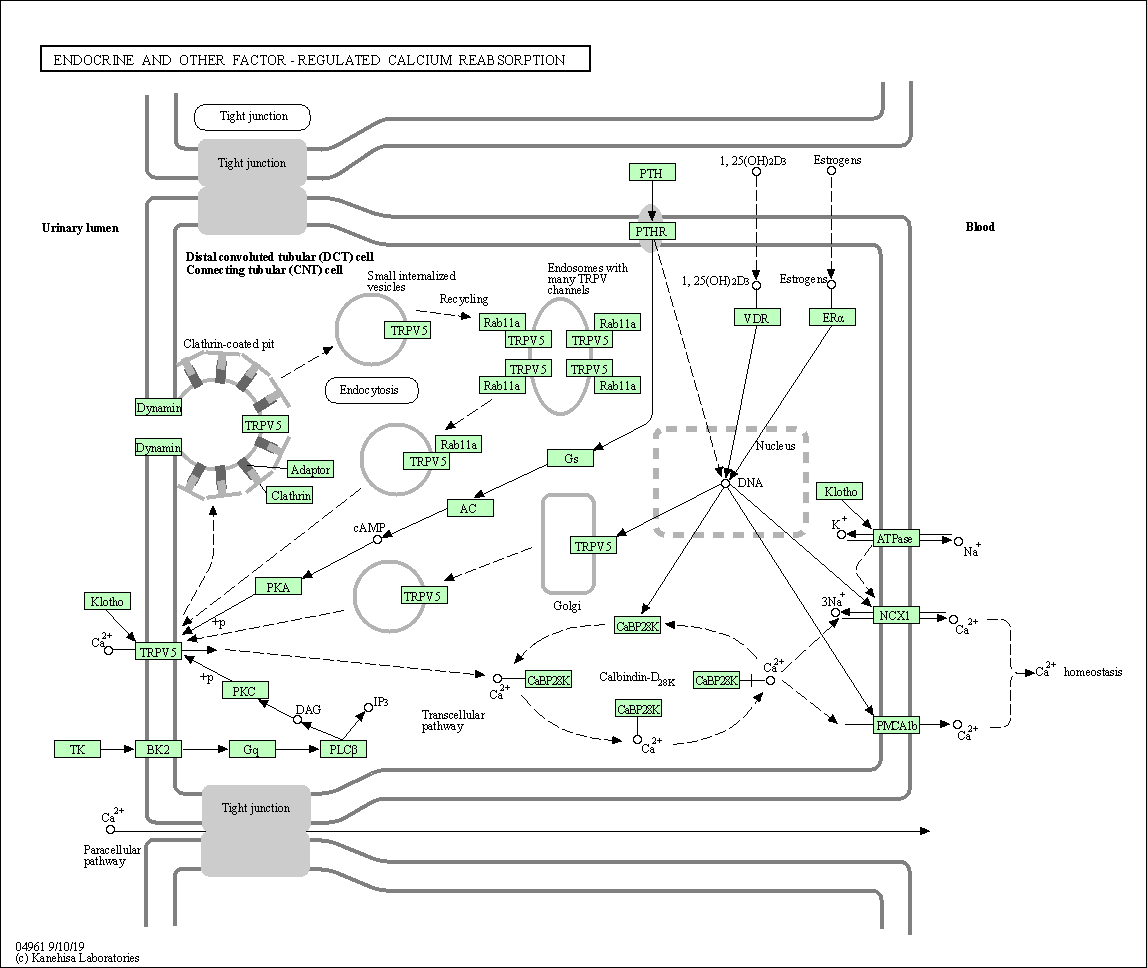
|
|||||||
| Vasopressin-regulated water reabsorption | hsa04962 |
Pathway Map 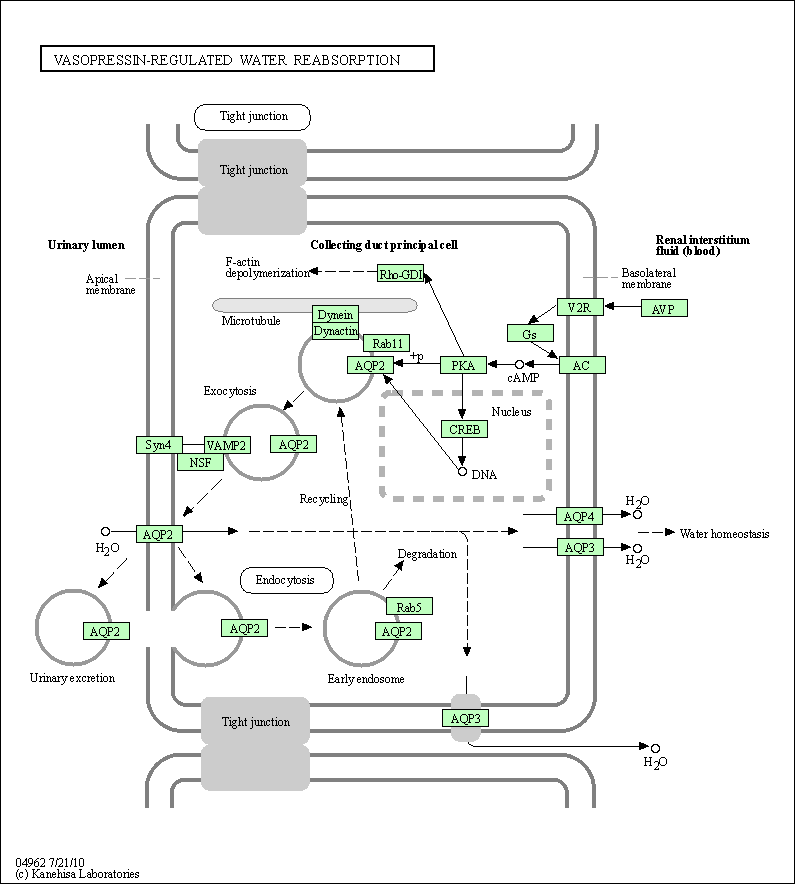
|
|||||||
| Long-term potentiation | hsa04720 |
Pathway Map 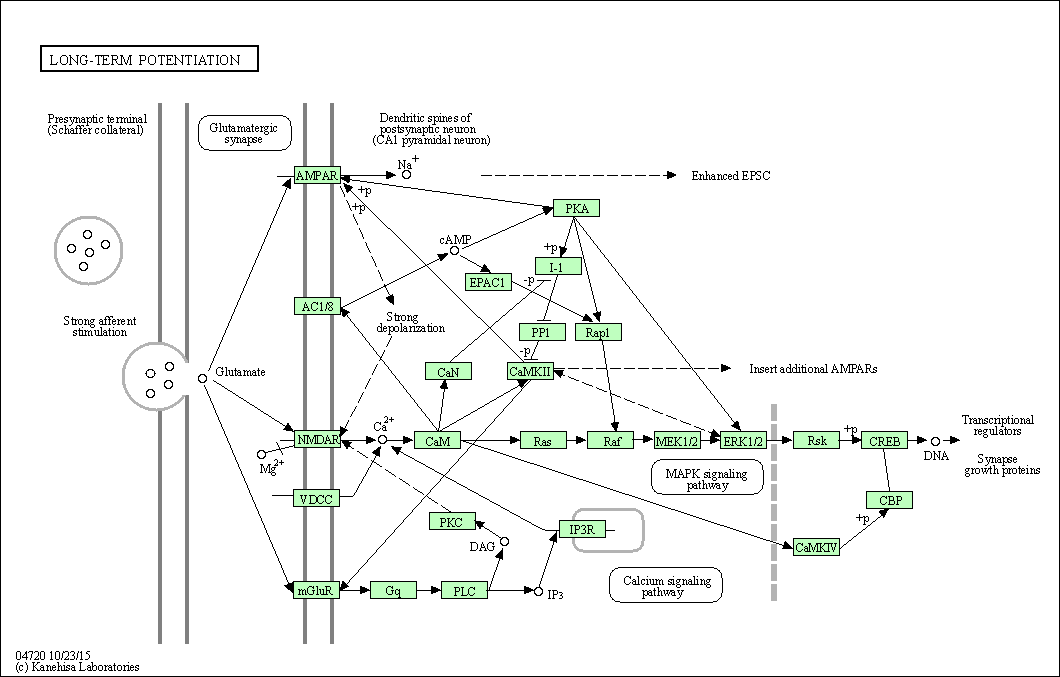
|
|||||||
| Retrograde endocannabinoid signaling | hsa04723 |
Pathway Map 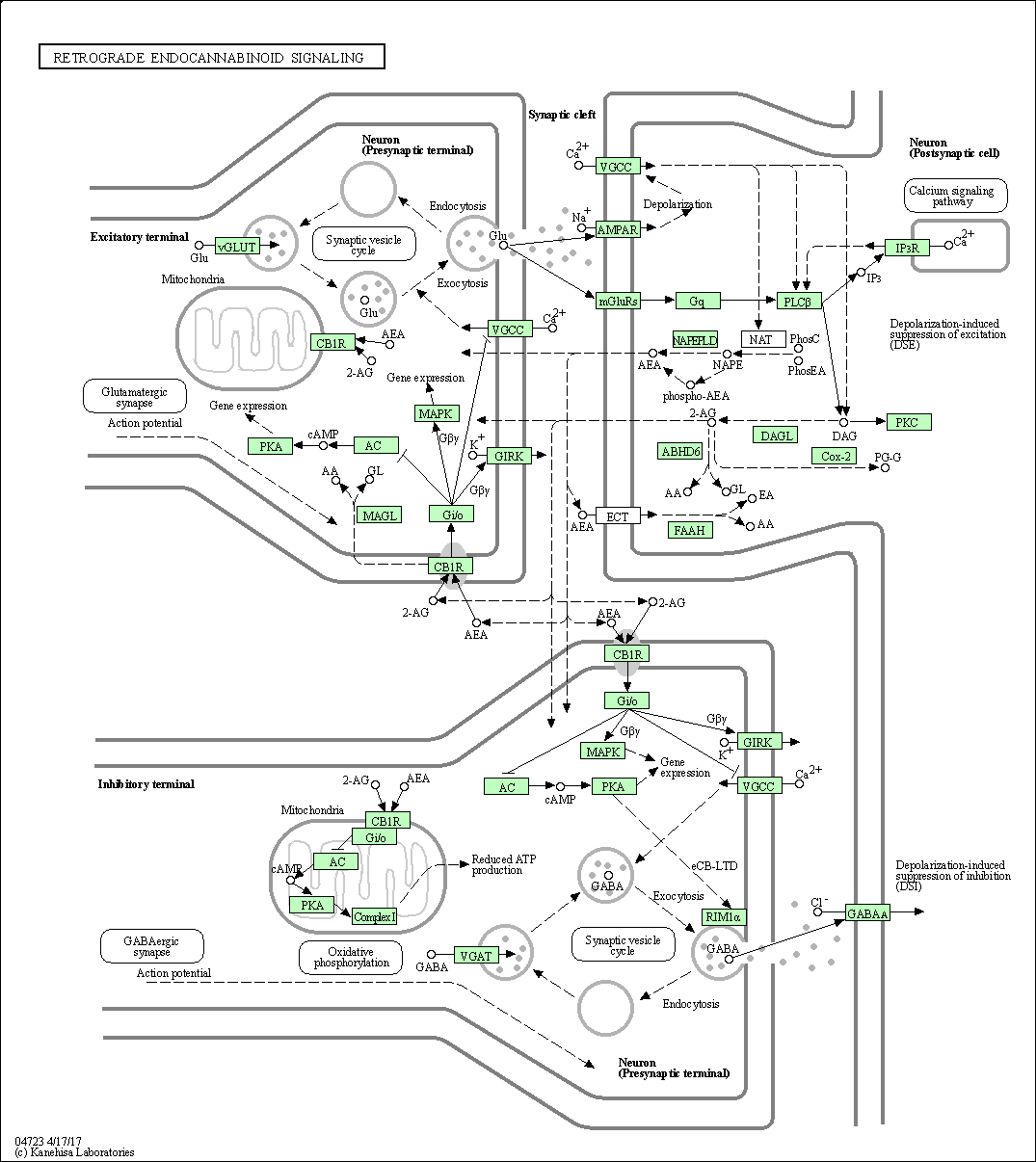
|
|||||||
| Glutamatergic synapse | hsa04724 |
Pathway Map 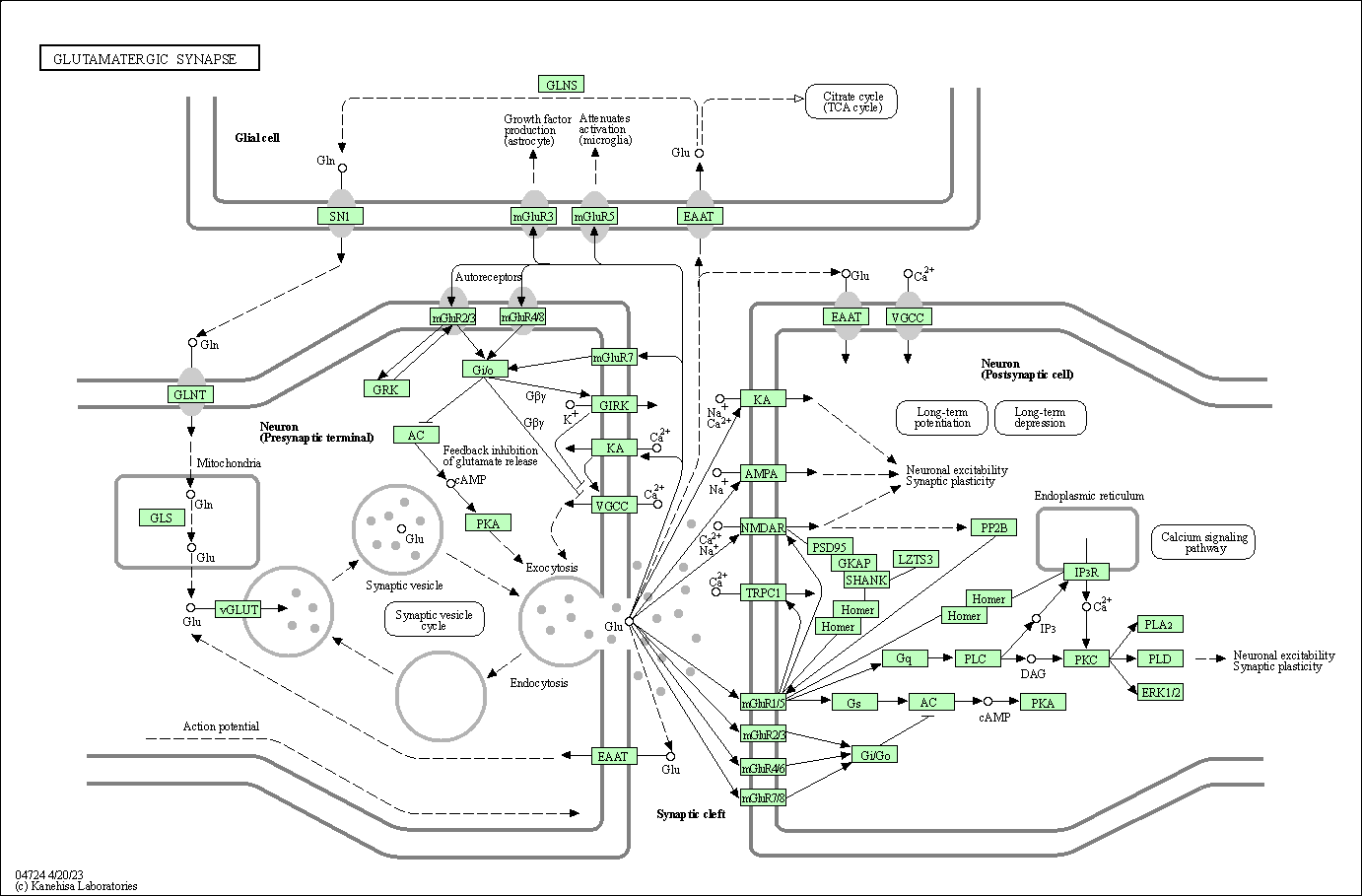
|
|||||||
| Cholinergic synapse | hsa04725 |
Pathway Map 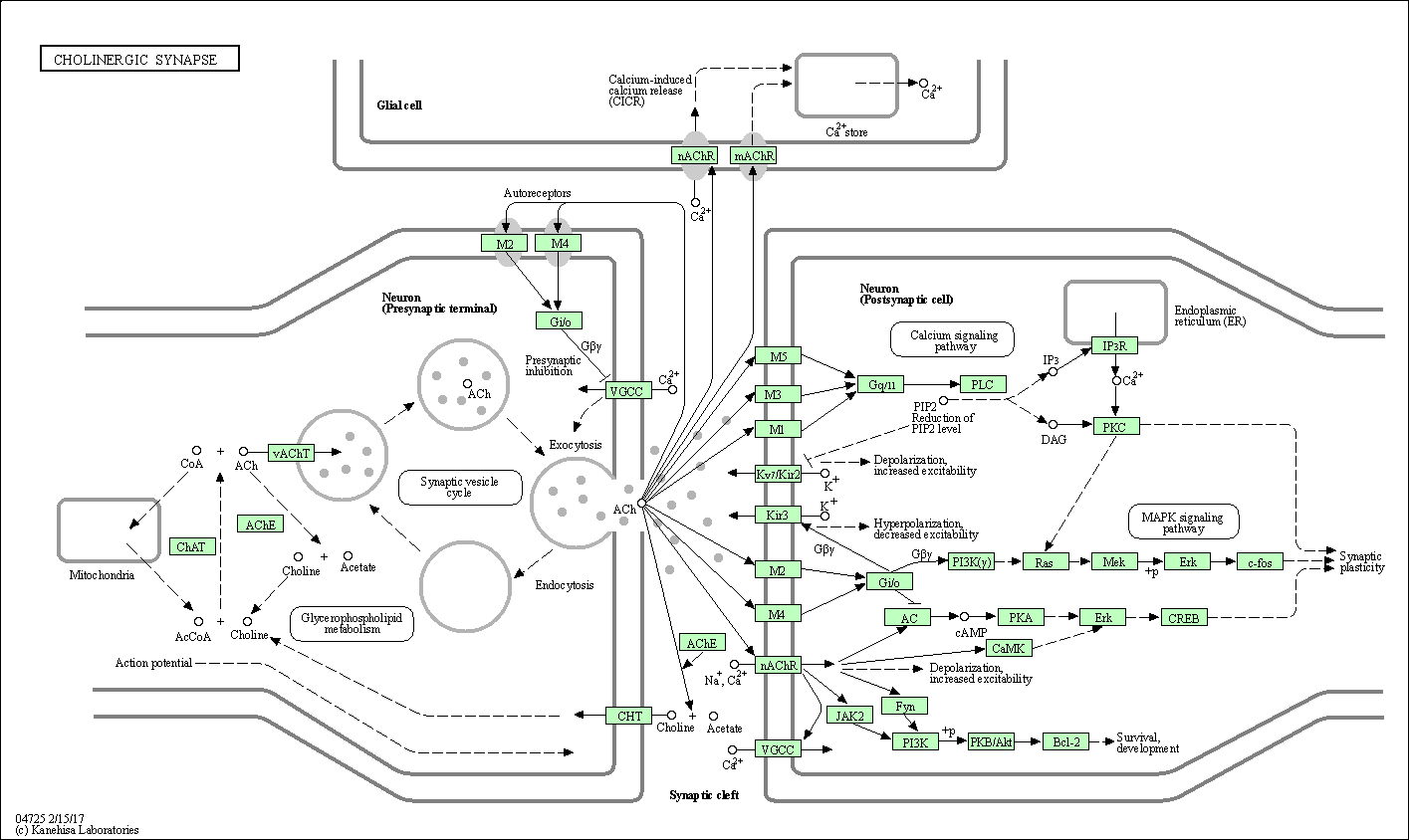
|
|||||||
| Serotonergic synapse | hsa04726 |
Pathway Map 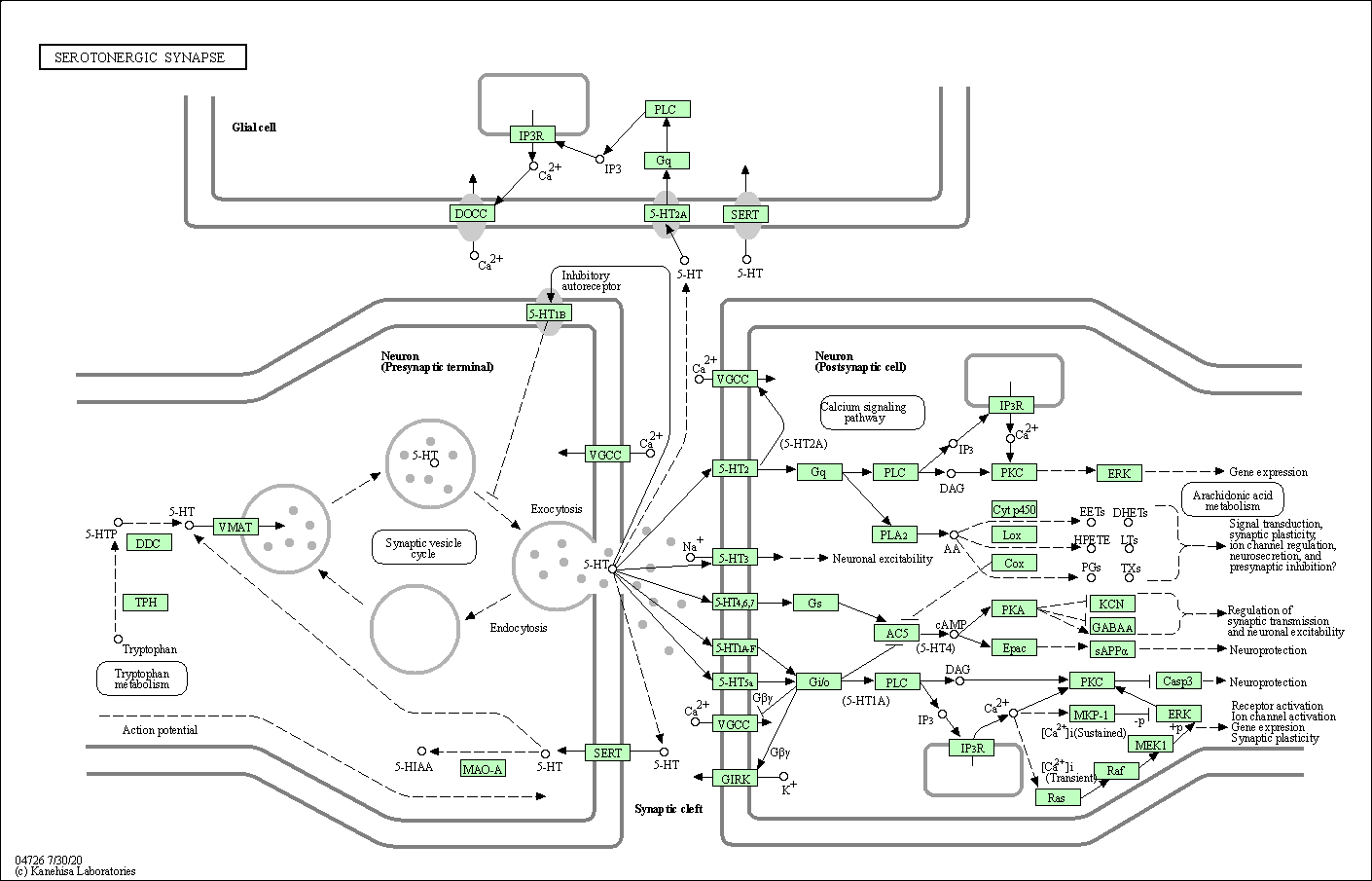
|
|||||||
| GABAergic synapse | hsa04727 |
Pathway Map 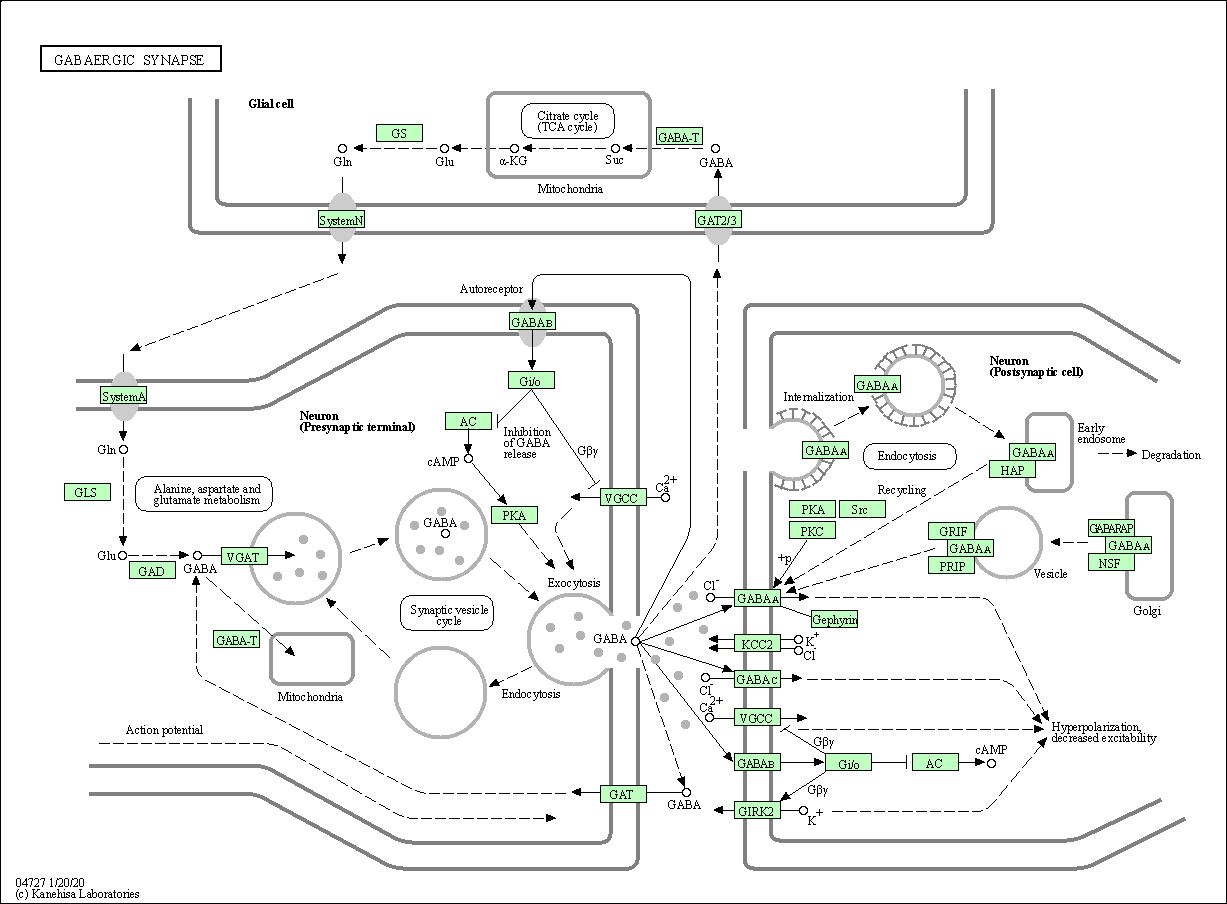
|
|||||||
| Dopaminergic synapse | hsa04728 |
Pathway Map 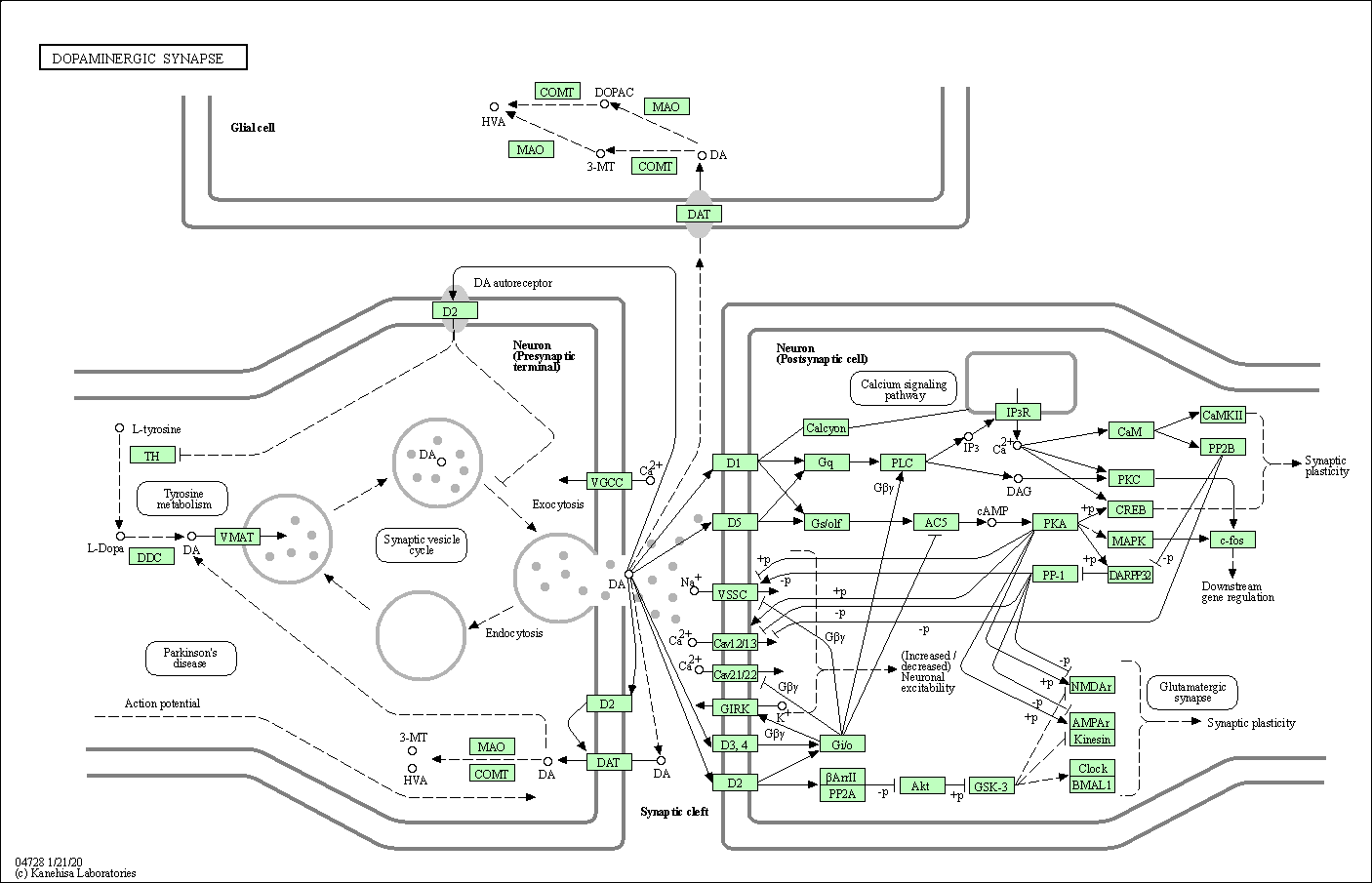
|
|||||||
| Parkinson disease | hsa05012 |
Pathway Map 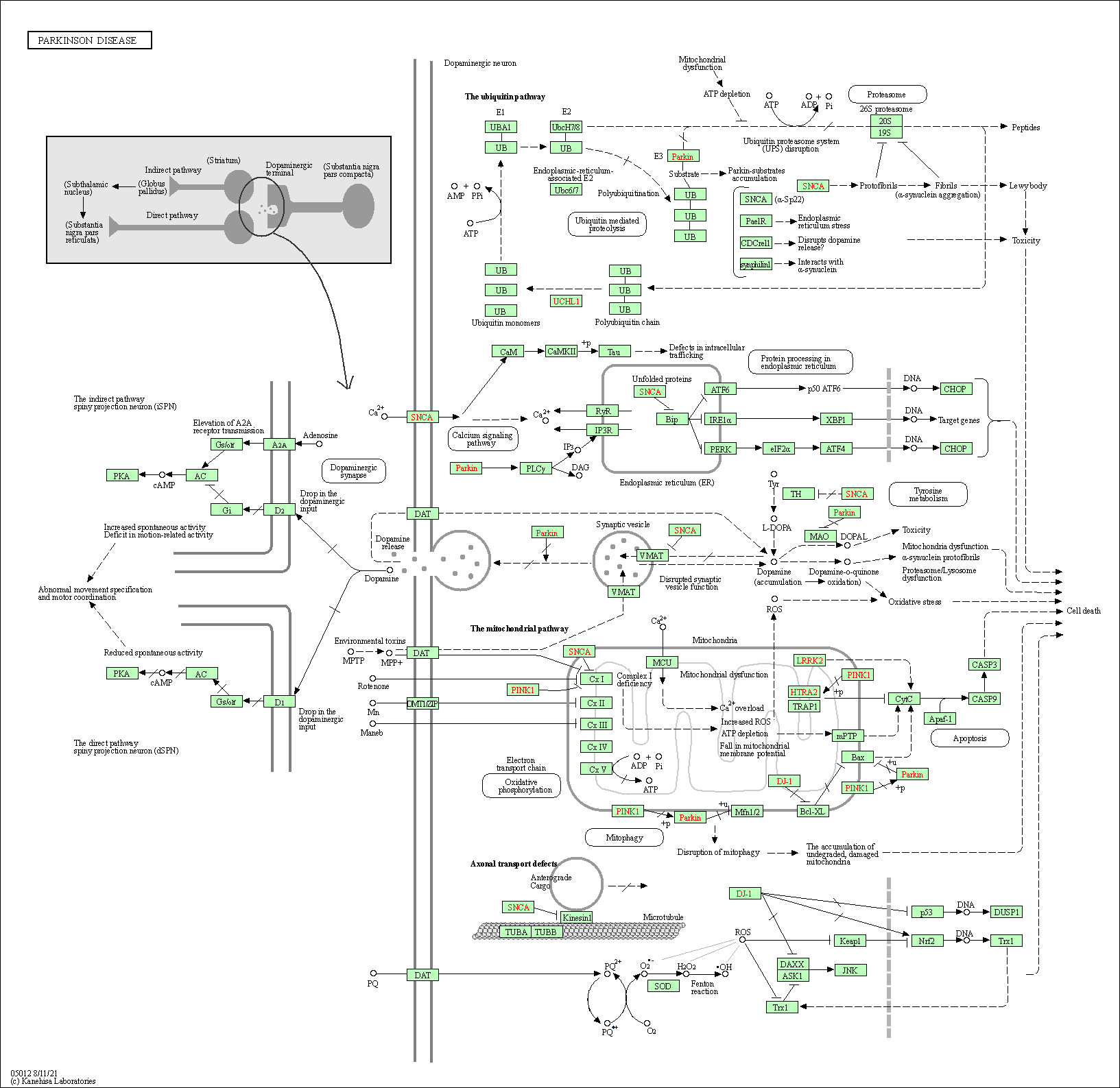
|
|||||||
| Prion disease | hsa05020 |
Pathway Map 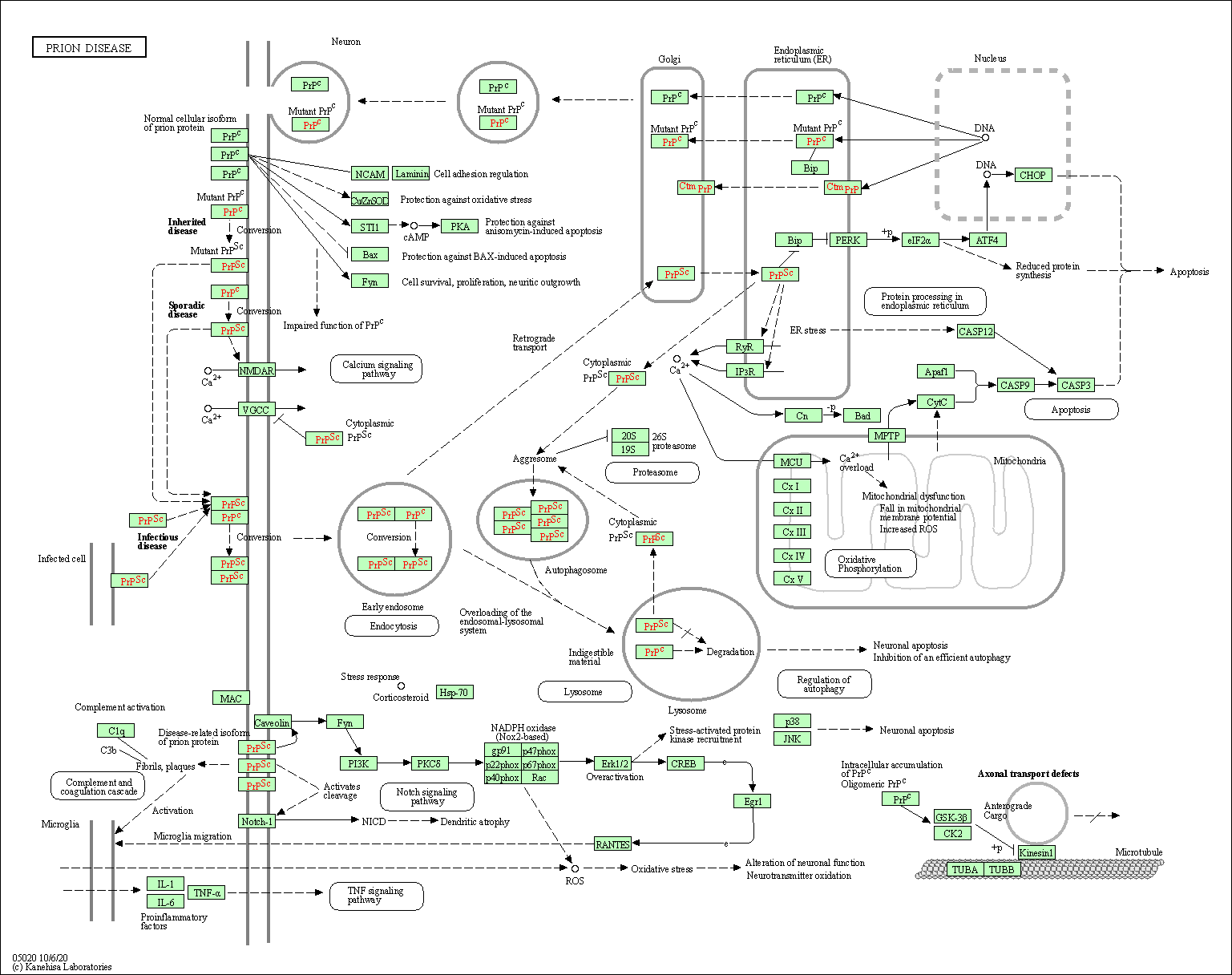
|
|||||||
| Olfactory transduction | hsa04740 |
Pathway Map 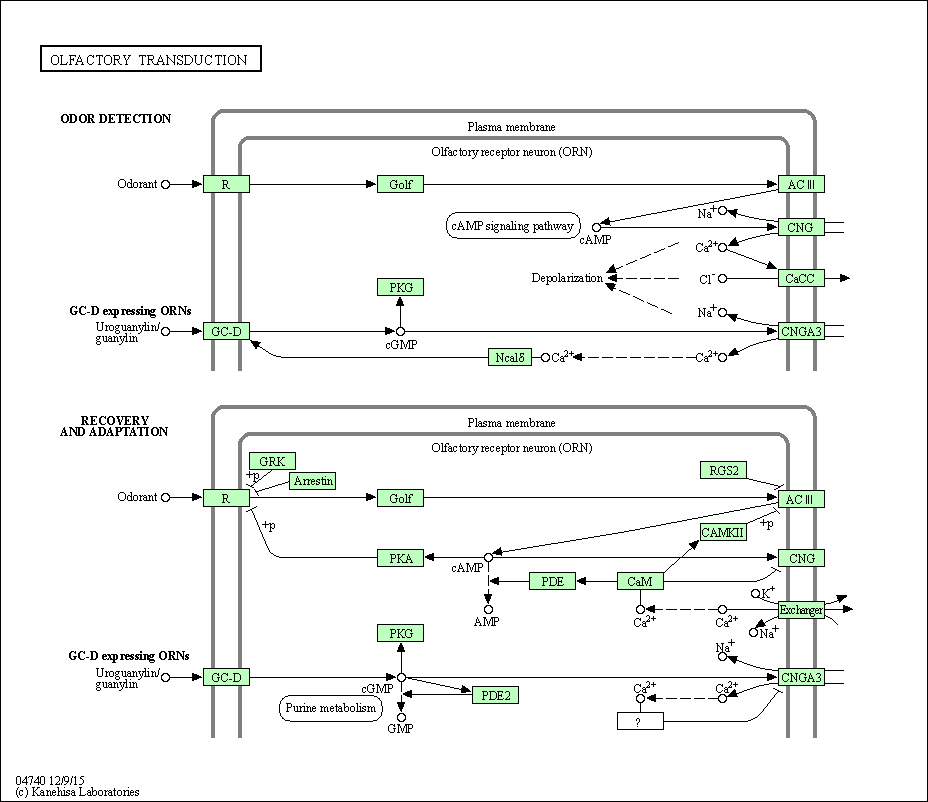
|
|||||||
| Taste transduction | hsa04742 |
Pathway Map 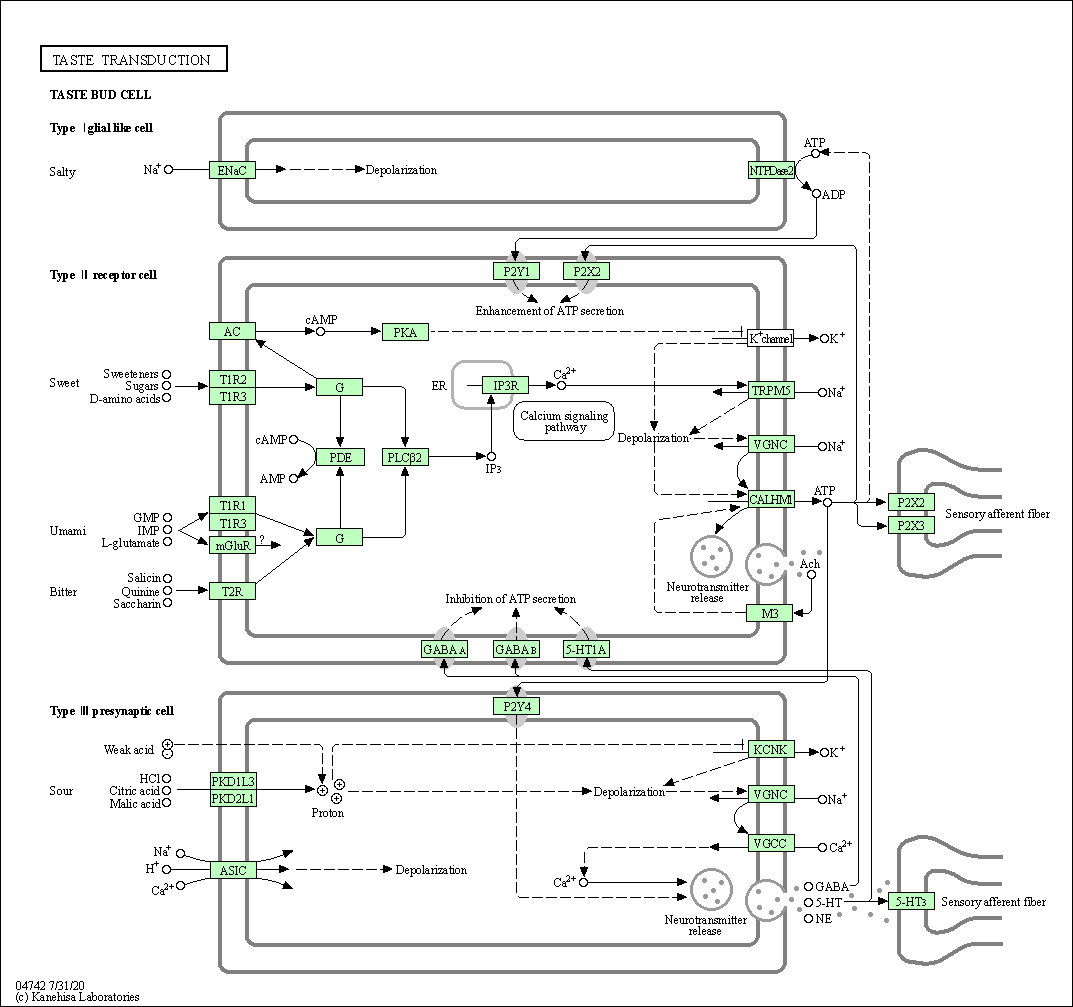
|
|||||||
| Inflammatory mediator regulation of TRP channels | hsa04750 |
Pathway Map 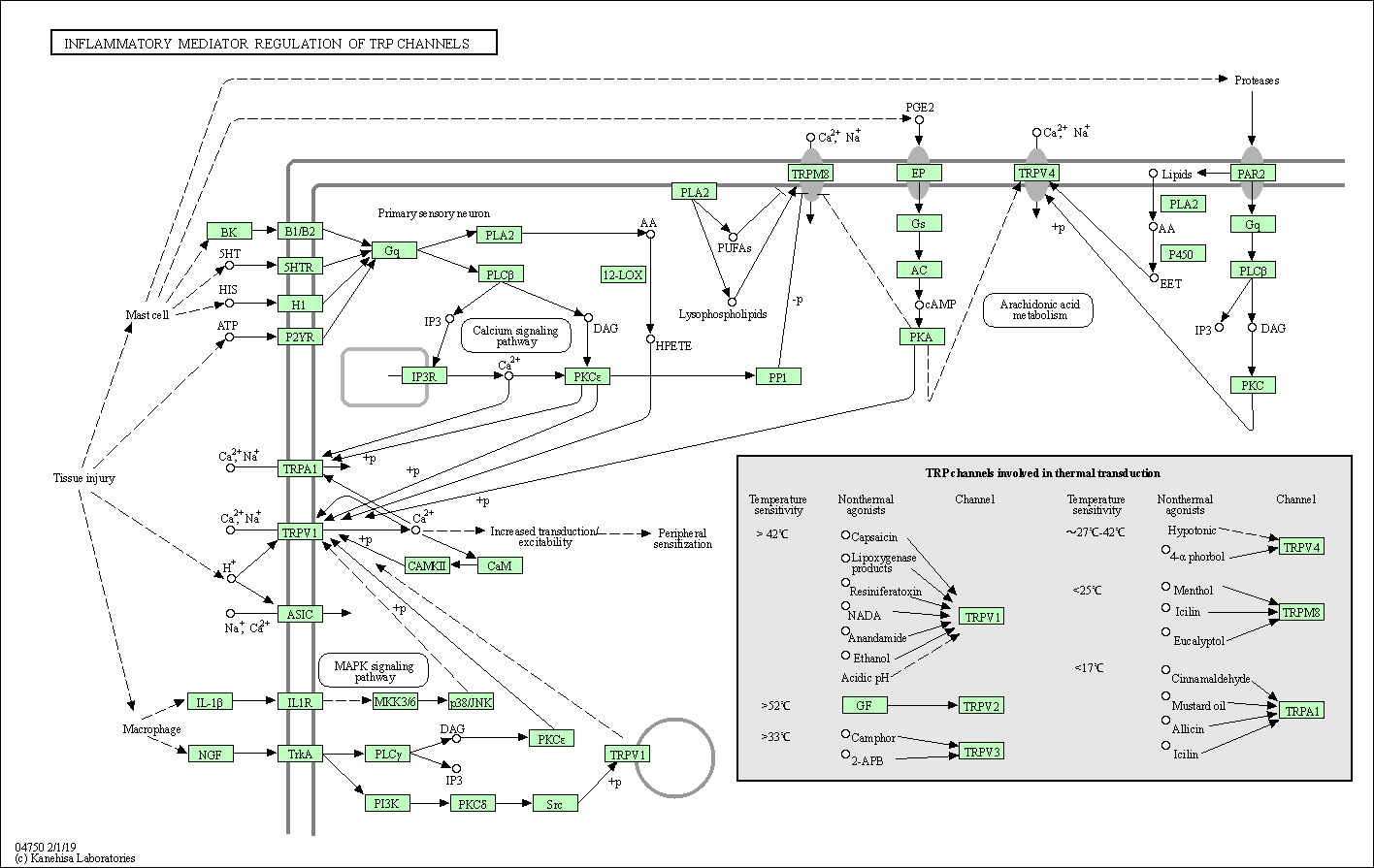
|
|||||||
| MAPK signaling pathway | hsa04010 |
Pathway Map 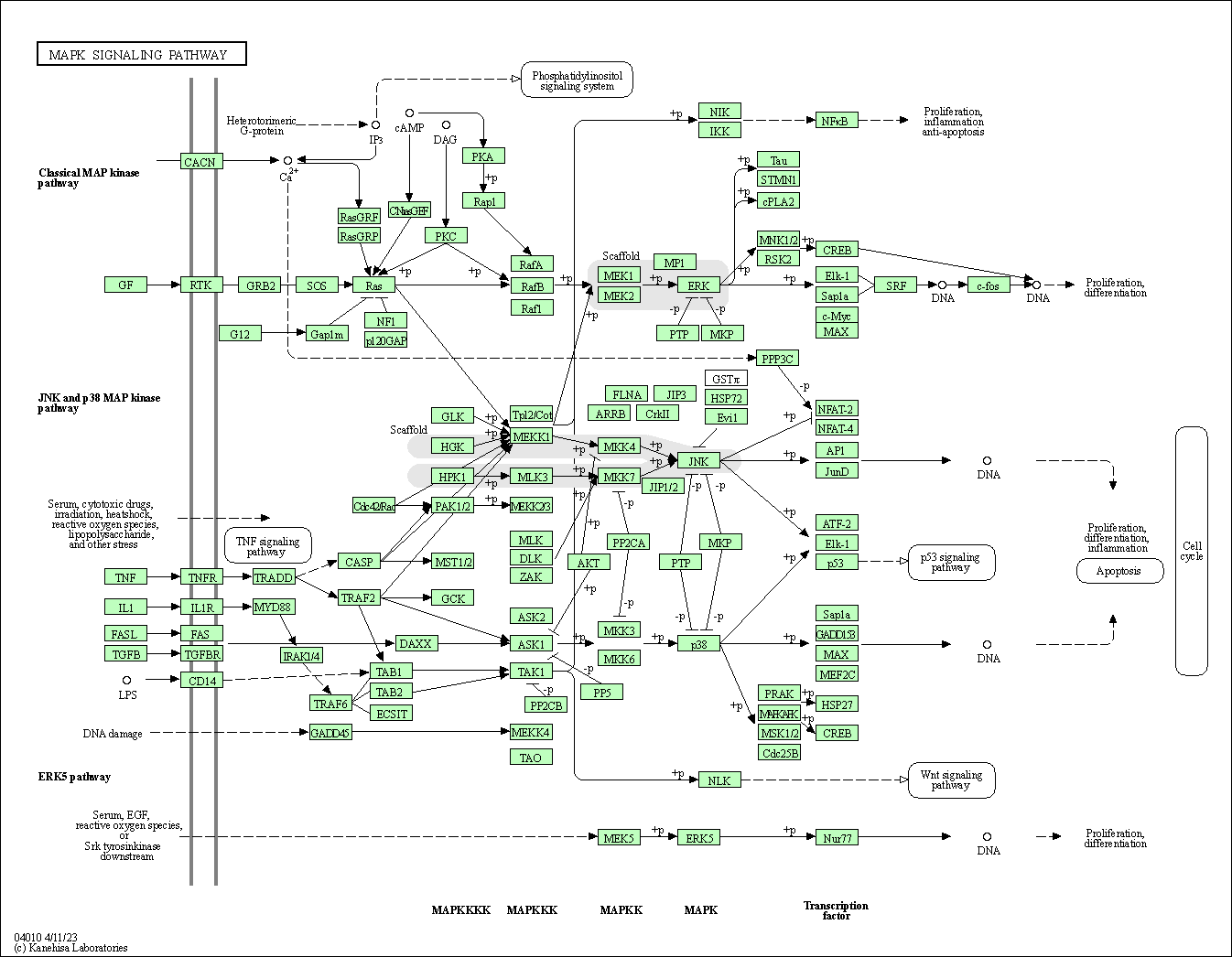
|
|||||||
| Ras signaling pathway | hsa04014 |
Pathway Map 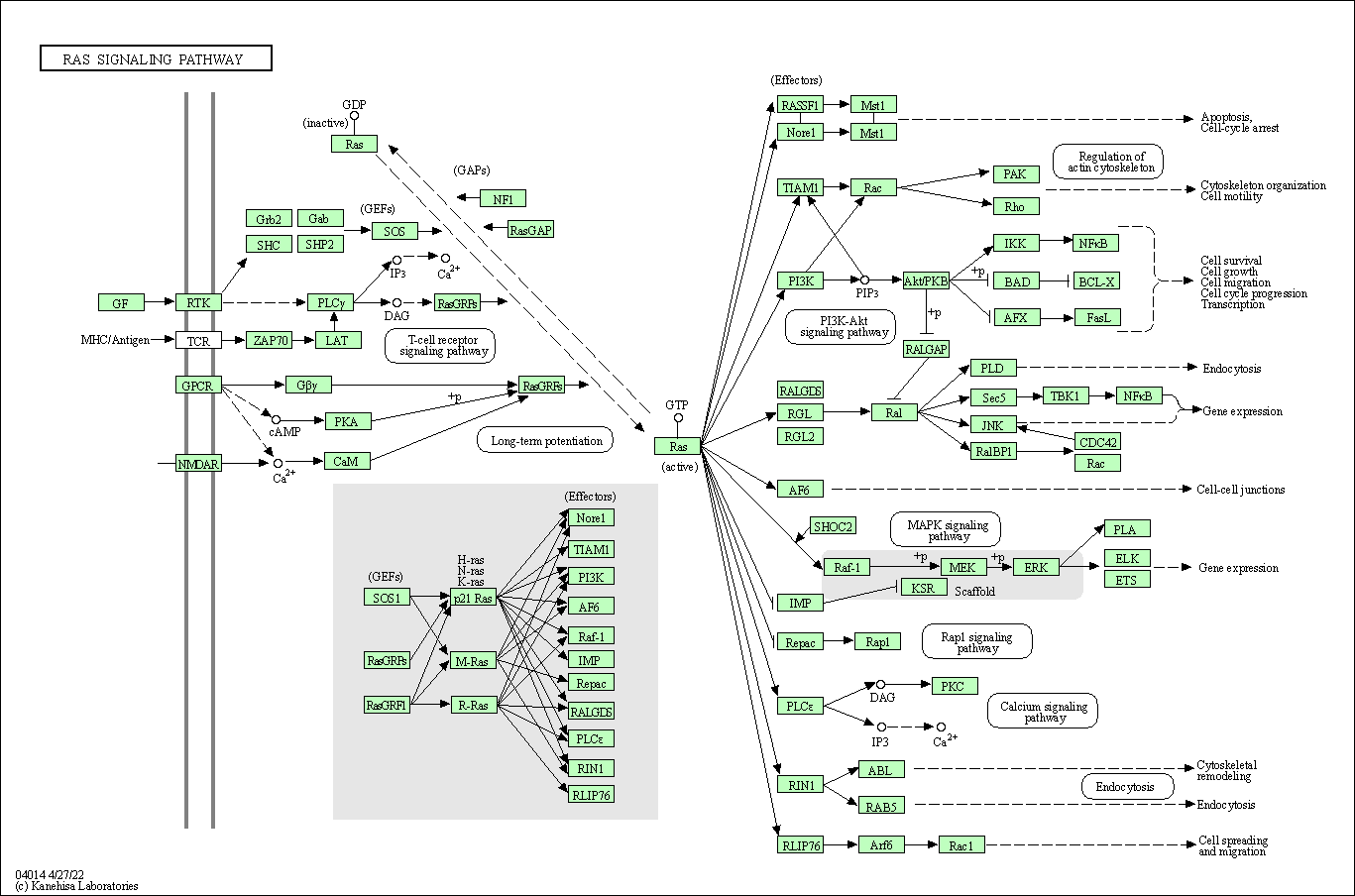
|
|||||||
| Calcium signaling pathway | hsa04020 |
Pathway Map 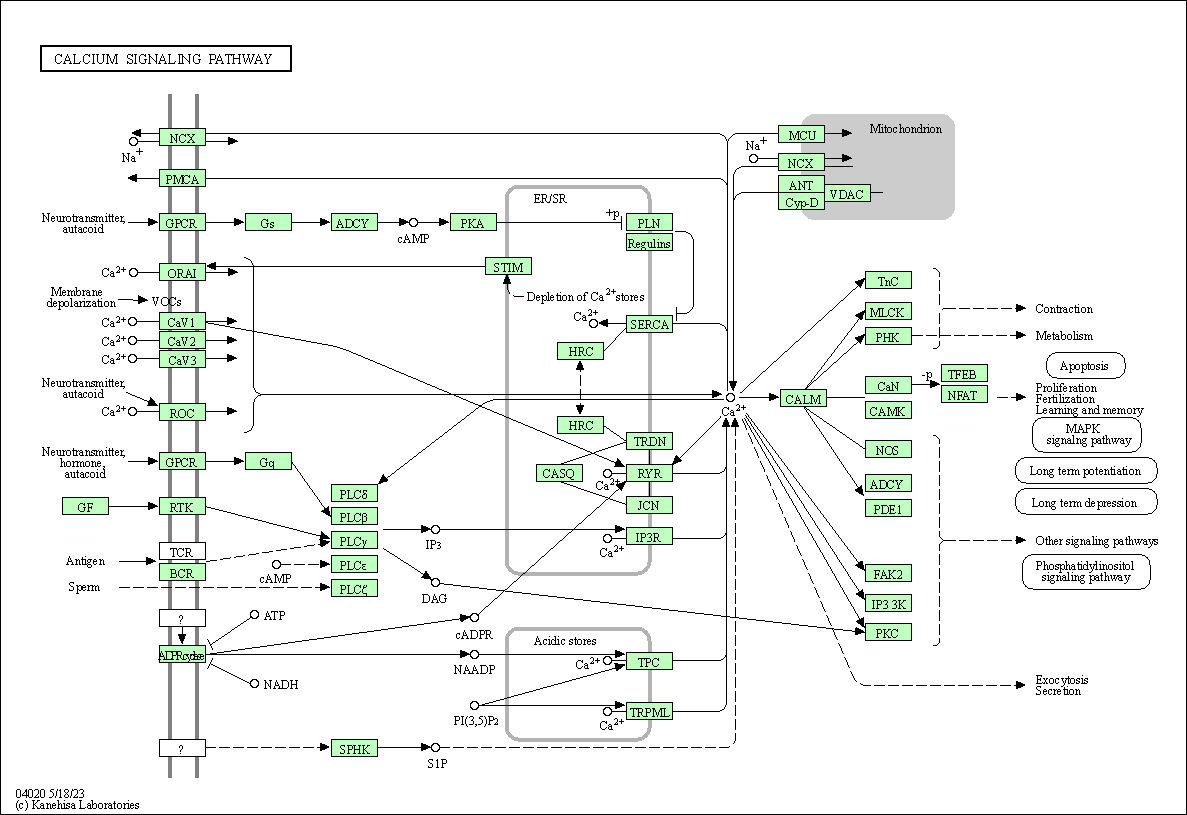
|
|||||||
| cAMP signaling pathway | hsa04024 |
Pathway Map 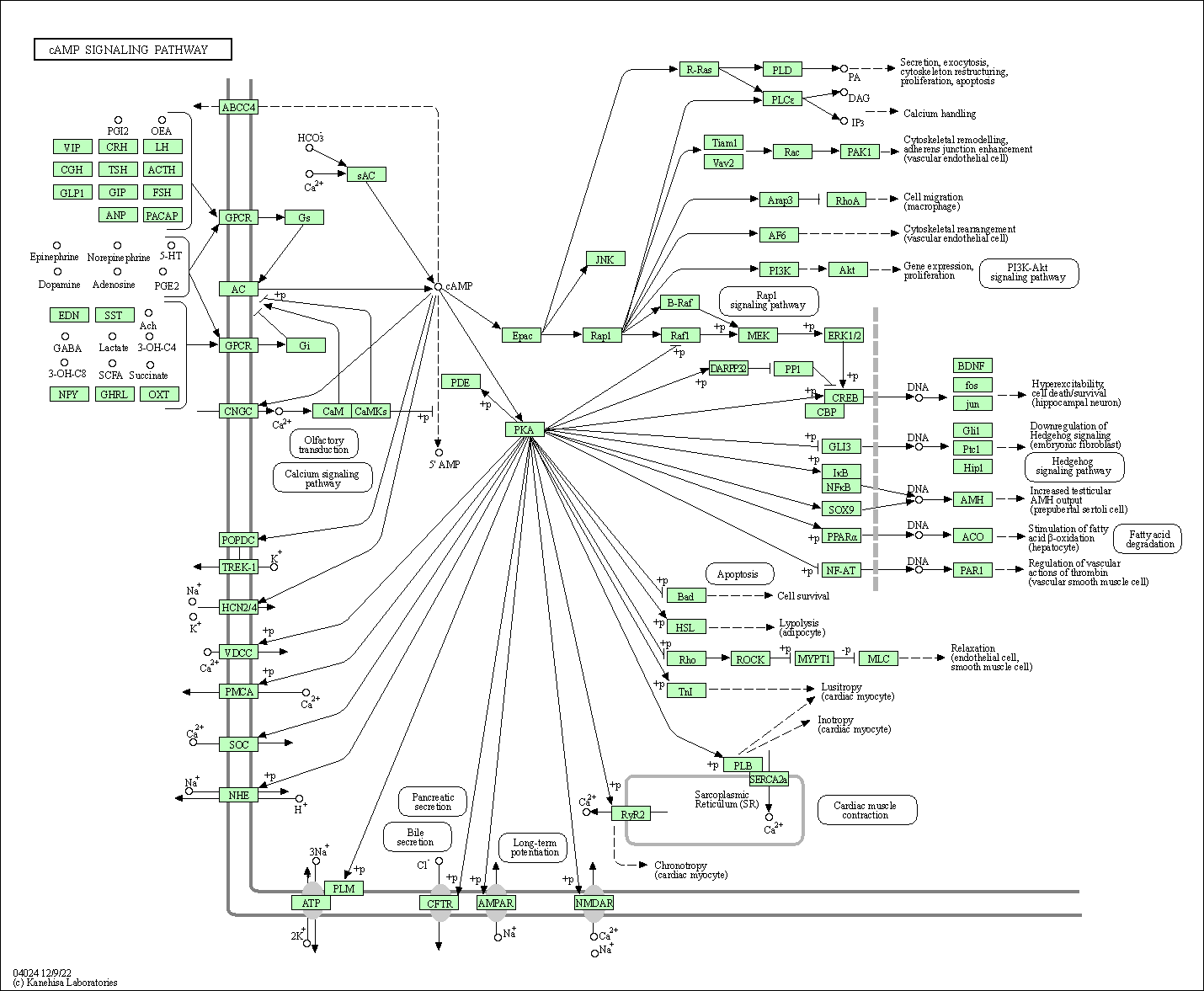
|
|||||||
| Wnt signaling pathway | hsa04310 |
Pathway Map 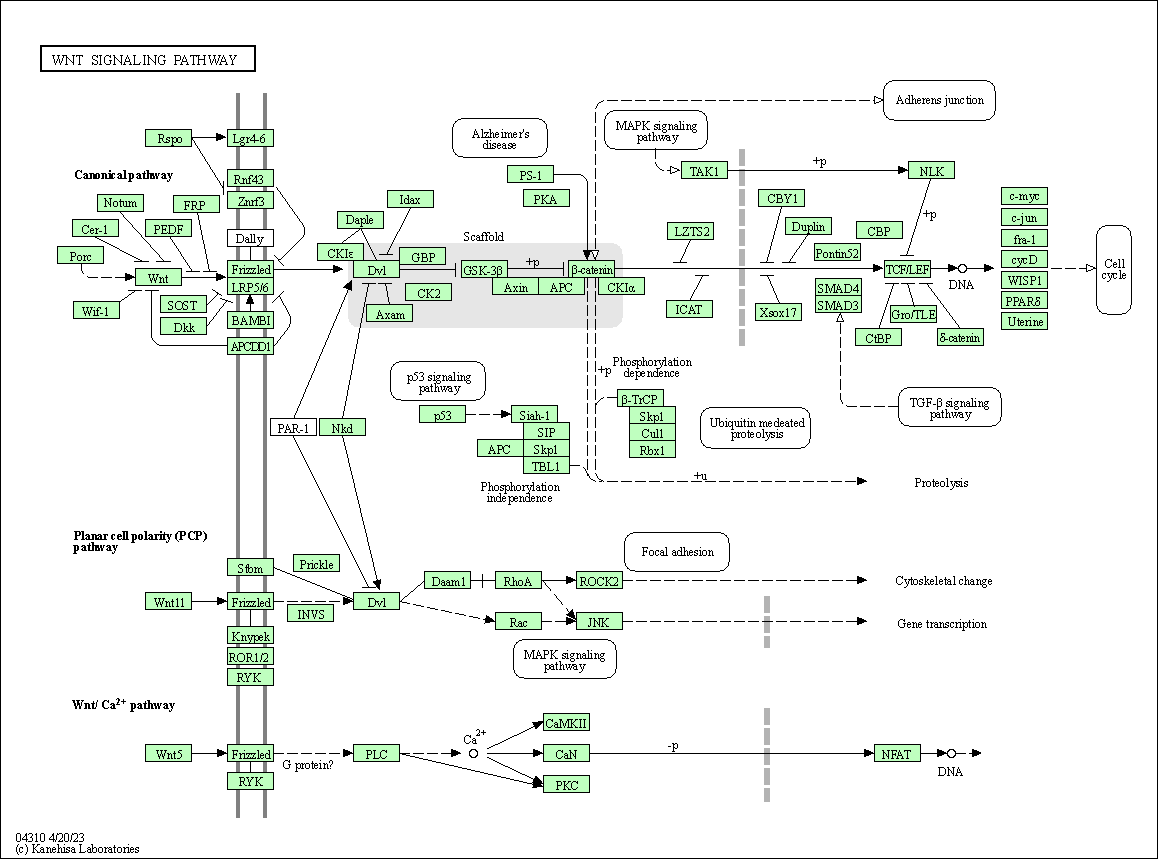
|
|||||||
| Hedgehog signaling pathway | hsa04340 |
Pathway Map 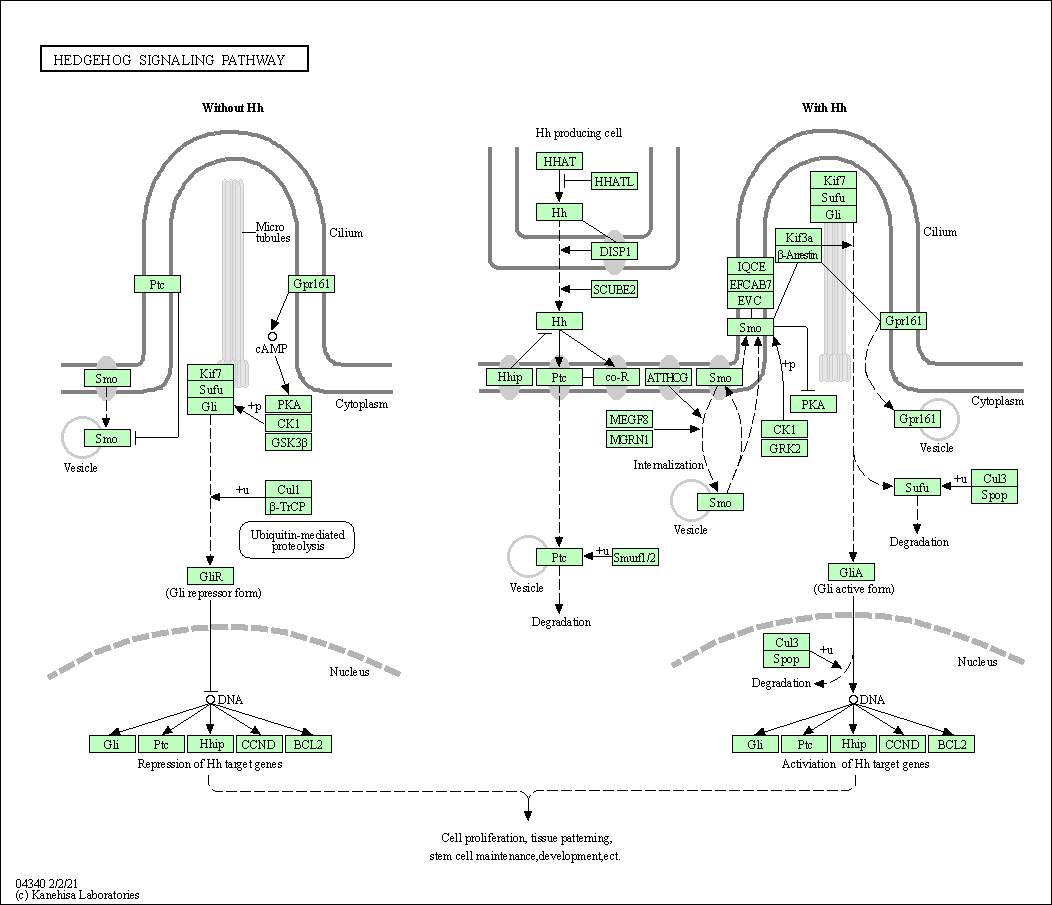
|
|||||||
| Apelin signaling pathway | hsa04371 |
Pathway Map 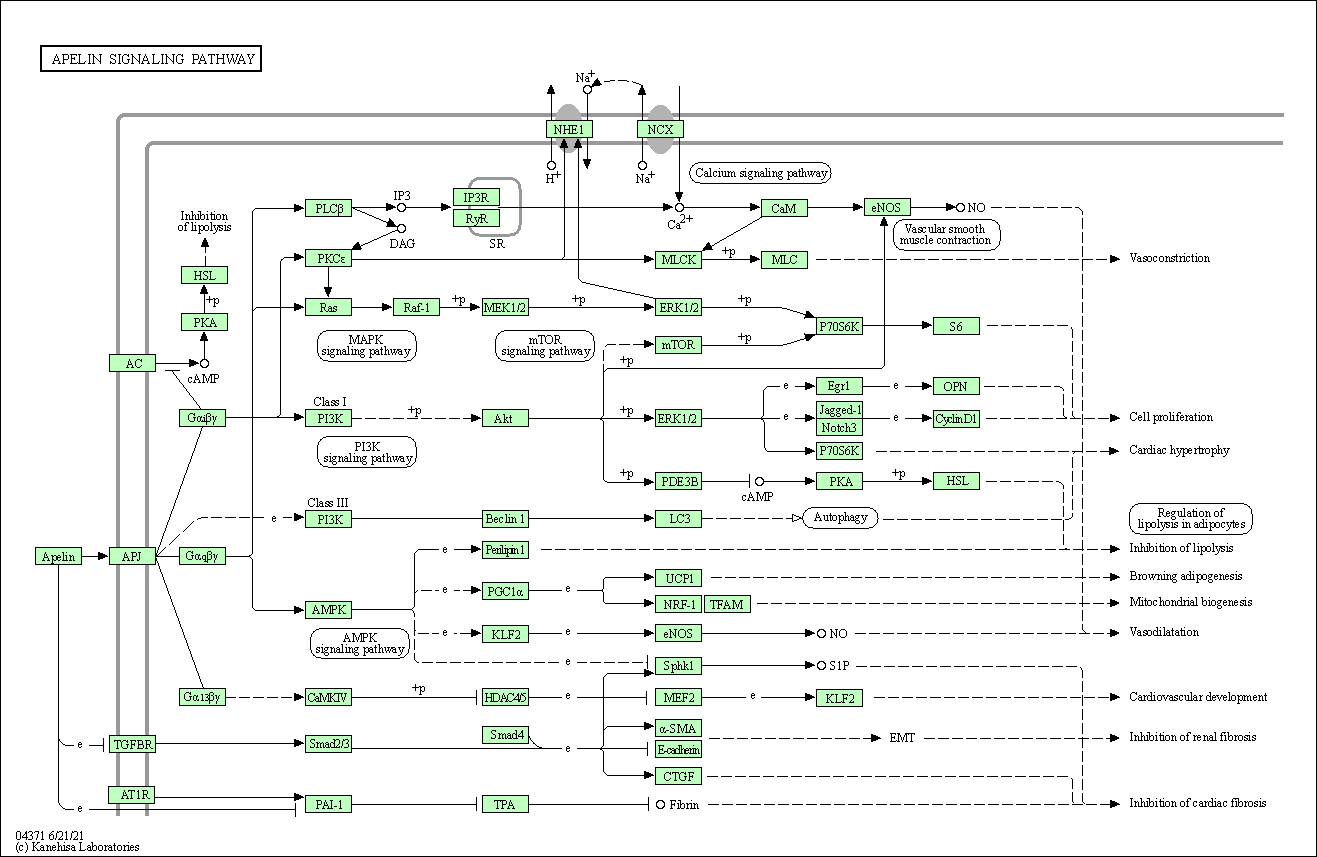
|
|||||||
| Cocaine addiction | hsa05030 |
Pathway Map 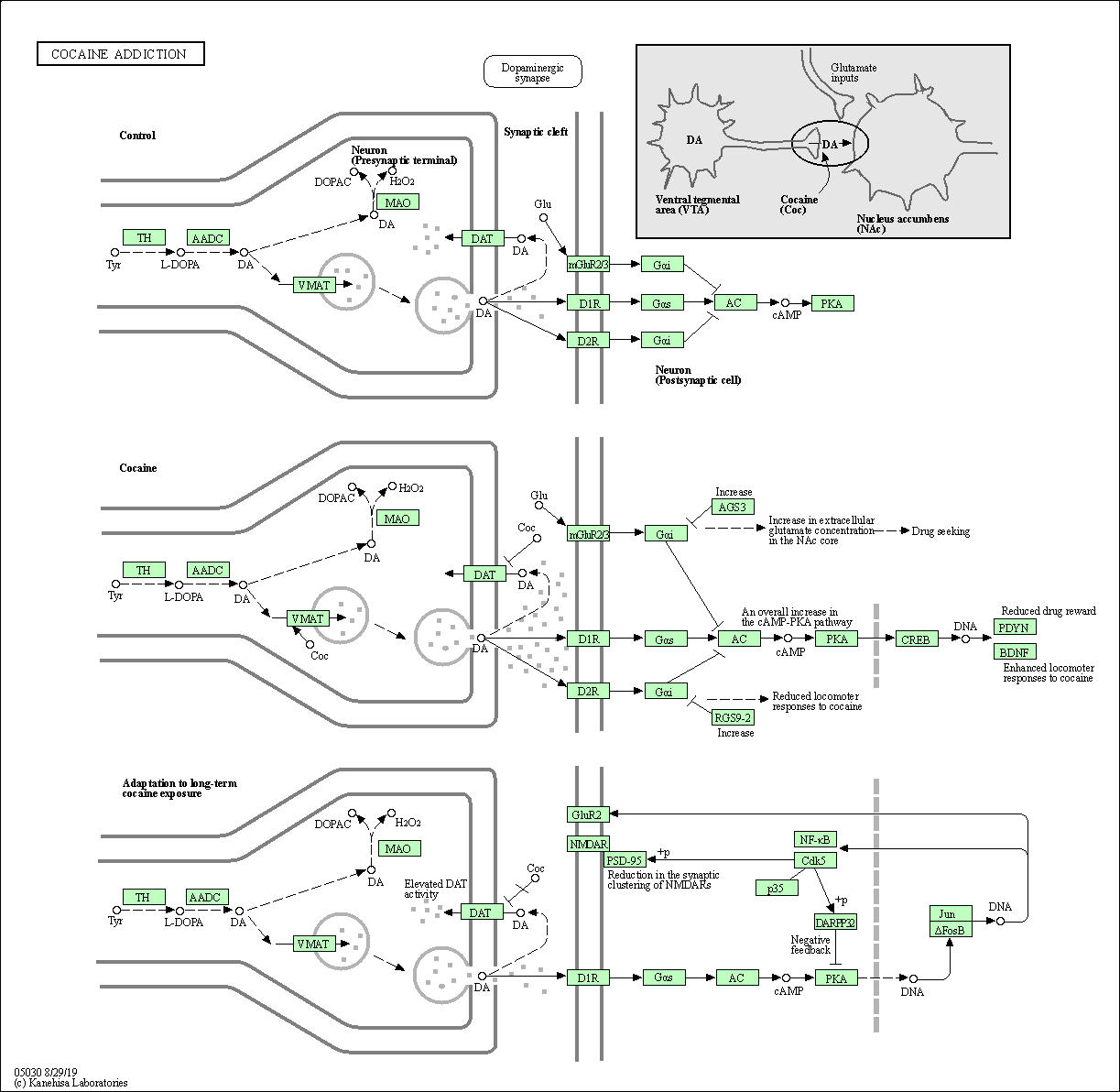
|
|||||||
| Amphetamine addiction | hsa05031 |
Pathway Map 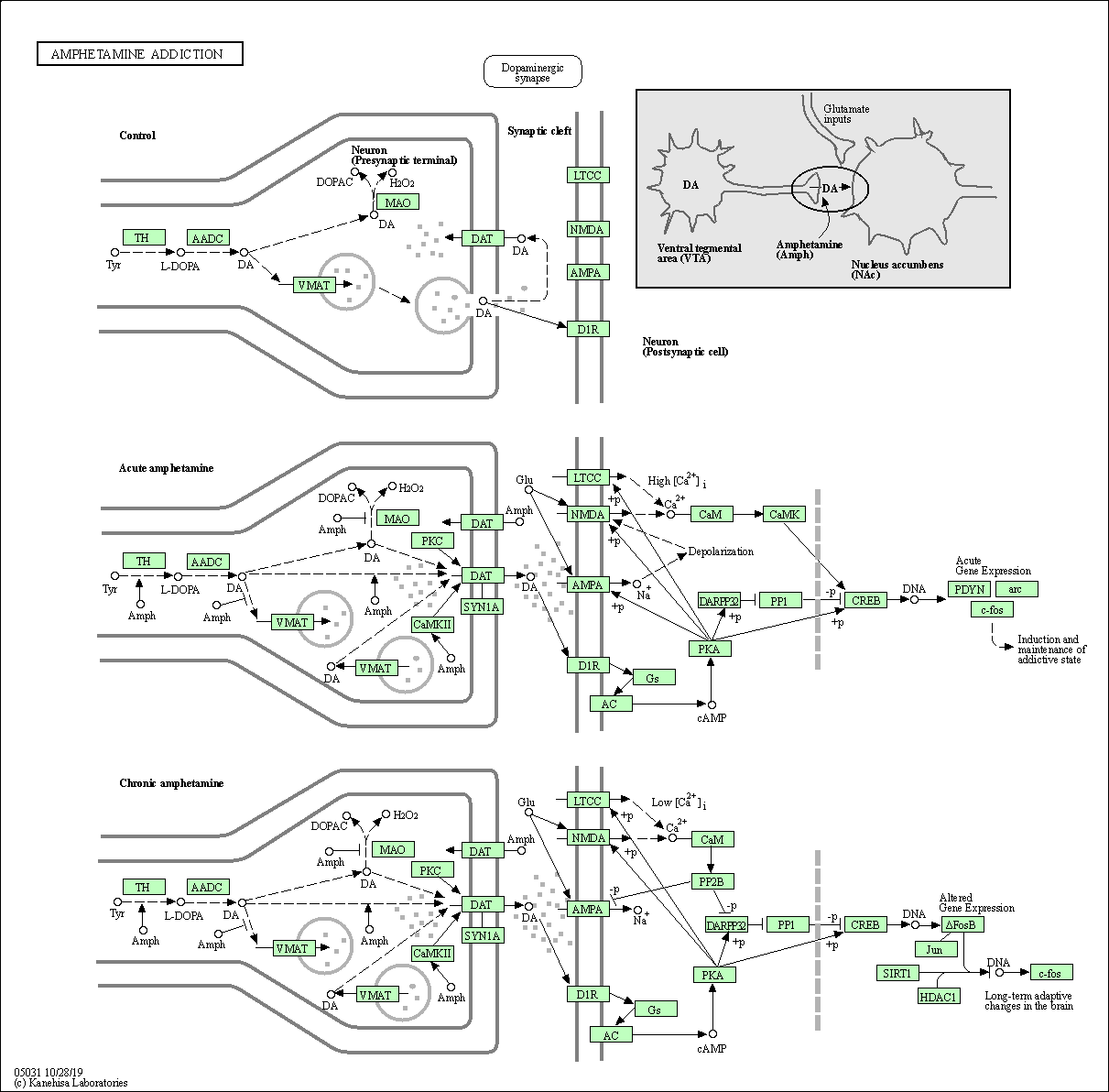
|
|||||||
| Morphine addiction | hsa05032 |
Pathway Map 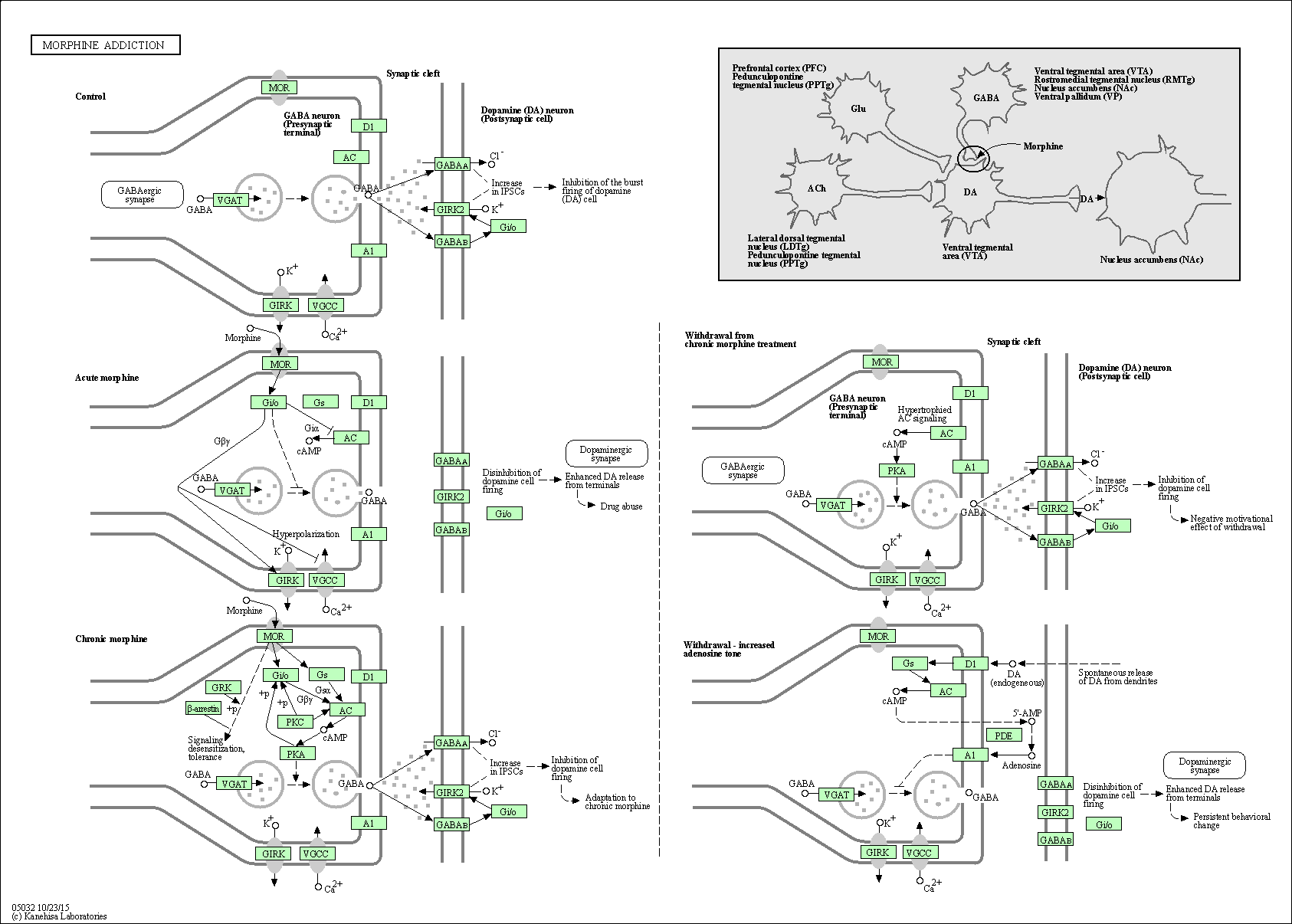
|
|||||||
| Alcoholism | hsa05034 |
Pathway Map 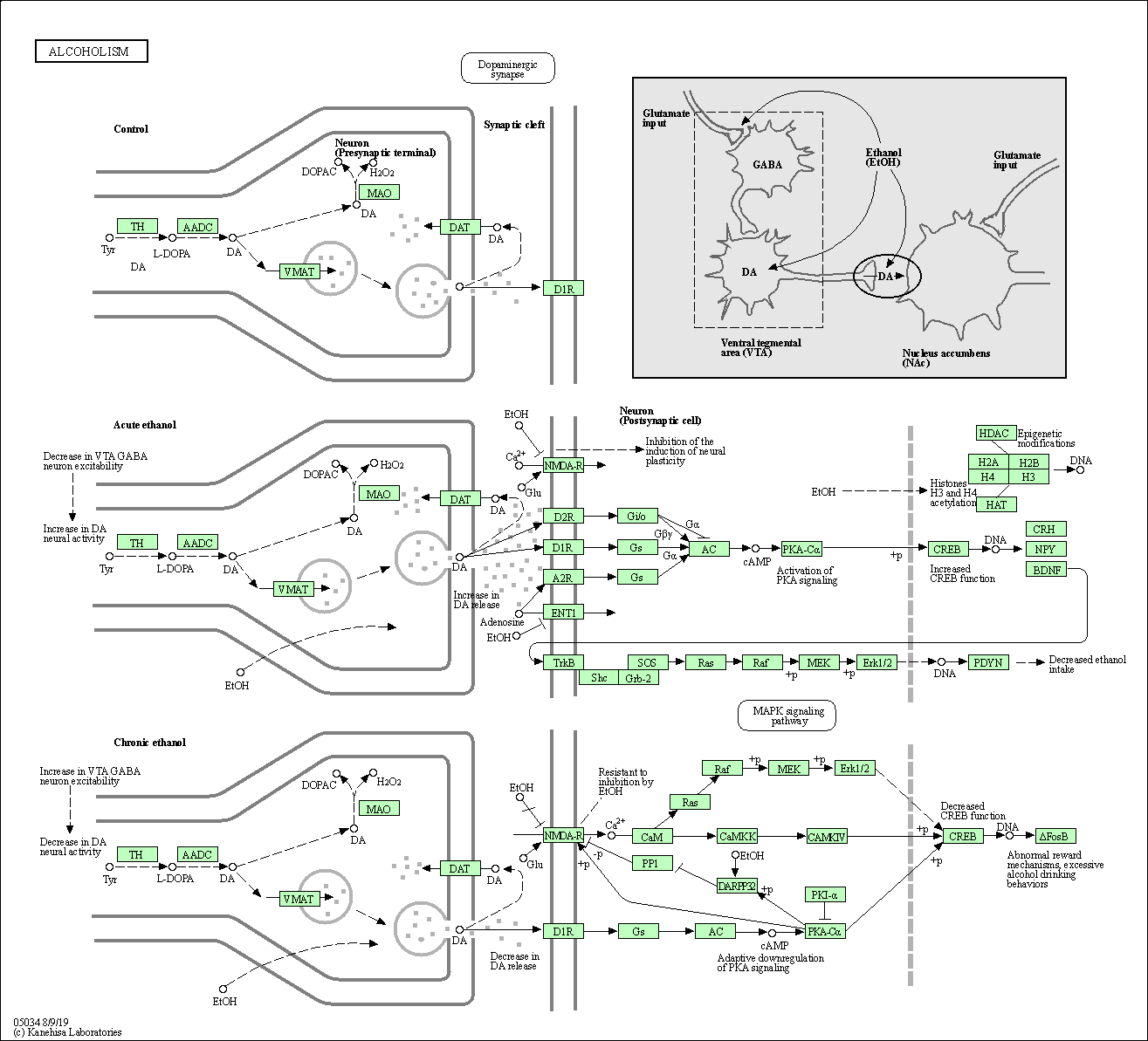
|
|||||||
| Autophagy - animal | hsa04140 |
Pathway Map 
|
|||||||
| 3D Structure |
|
||||||||
Full List of Virus RNA Interacting with This Protien
| Virus Name | Binding Region | RNA Info | Detection Method | Infection Cell | Cell ID | Cell Originated Tissue | Infection Time | Interaction Score 1 | Interaction Score 2 |
|---|---|---|---|---|---|---|---|---|---|
| Encephalomyocarditis virus | 5'UTR - 3'UTR | RNA Info | Comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS) | HuH-7.5 Cells (Human hepatocellular carcinoma cell) | . | Liver | . | Z-score = -1.164598859 | FZR = 1 |
| Influenza A virus | 5'UTR - 3'UTR | RNA Info | Comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS) | HuH-7.5 Cells (Human hepatocellular carcinoma cell) | . | Liver | . | Z-score = 0.17733093 | FZR = 1 |
Potential Drug(s) that Targets This Protein
Protein Sequence Information
|
MGNAATAKKGSEVESVKEFLAKAKEDFLKKWENPTQNNAGLEDFERKKTLGTGSFGRVMLVKHKATEQYYAMKILDKQKVVKLKQIEHTLNEKRILQAVNFPFLVRLEYAFKDNSNLYMVMEYVPGGEMFSHLRRIGRFSEPHARFYAAQIVLTFEYLHSLDLIYRDLKPENLLIDHQGYIQVTDFGFAKRVKGRTWTLCGTPEYLAPEIILSKGYNKAVDWWALGVLIYEMAAGYPPFFADQPIQIYEKIVSGKVRFPSHFSSDLKDLLRNLLQVDLTKRFGNLKNGVSDIKTHKWFATTDWIAIYQRKVEAPFIPKFRGSGDTSNFDDYEEEDIRVSITEKCAKEFGEF
Click to Show/Hide
|
