Host Protein General Information
| Protein Name |
MAP kinase kinase 2
|
Gene Name |
MAP2K2
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Host Species |
Homo sapiens
|
Uniprot Entry Name |
MP2K2_HUMAN
|
||||||
| Protein Families |
Protein kinase superfamily, STE Ser/Thr protein kinase family, MAP kinase kinase subfamily
|
||||||||
| EC Number |
2.7.12.2
|
||||||||
| Subcellular Location |
Cytoplasm
|
||||||||
| External Link | |||||||||
| NCBI Gene ID | |||||||||
| Uniprot ID | |||||||||
| Ensembl ID | |||||||||
| HGNC ID | |||||||||
| Related KEGG Pathway | |||||||||
| Neutrophil extracellular trap formation | hsa04613 |
Pathway Map 
|
|||||||
| Toll-like receptor signaling pathway | hsa04620 |
Pathway Map 
|
|||||||
| Natural killer cell mediated cytotoxicity | hsa04650 |
Pathway Map 
|
|||||||
| T cell receptor signaling pathway | hsa04660 |
Pathway Map 
|
|||||||
| B cell receptor signaling pathway | hsa04662 |
Pathway Map 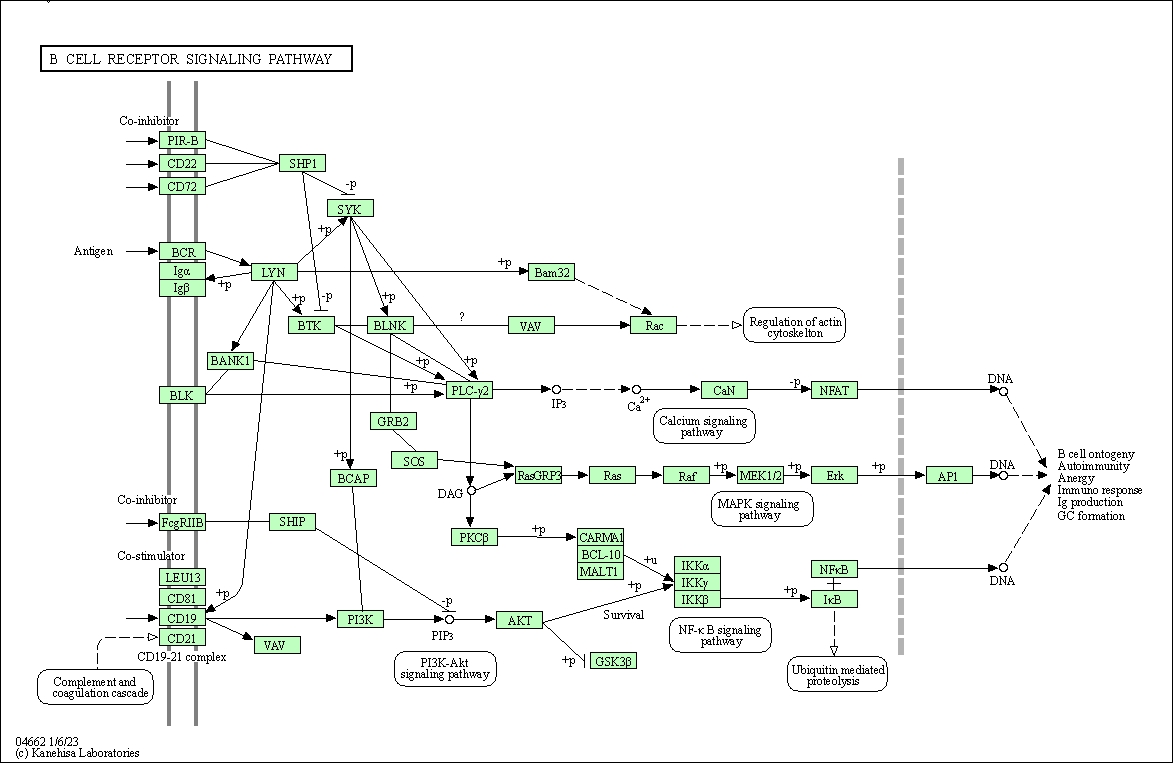
|
|||||||
| Fc epsilon RI signaling pathway | hsa04664 |
Pathway Map 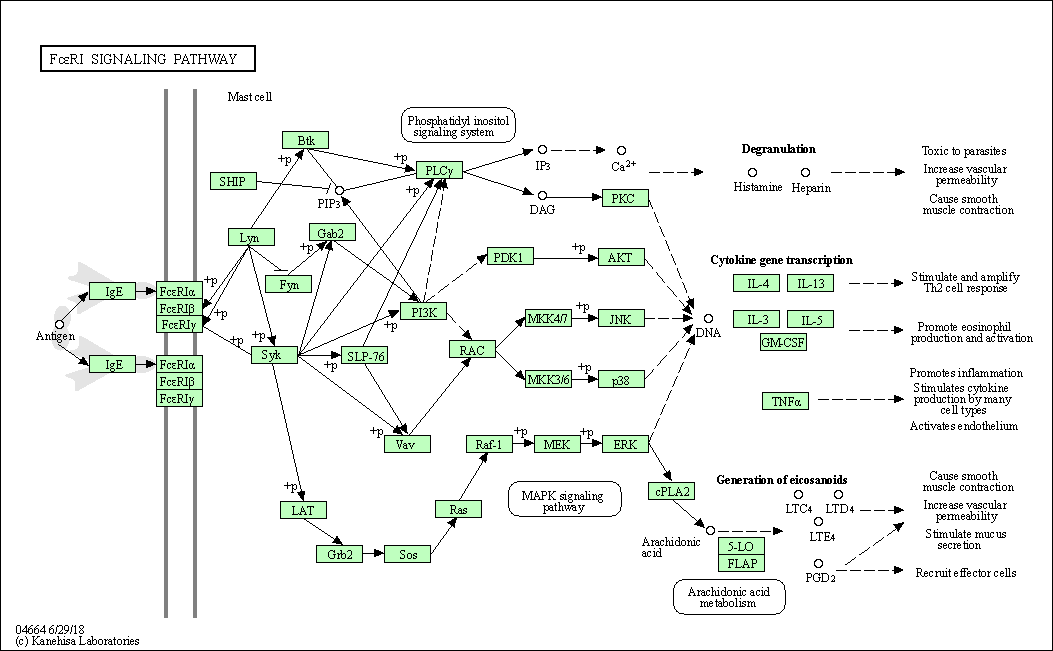
|
|||||||
| Salmonella infection | hsa05132 |
Pathway Map 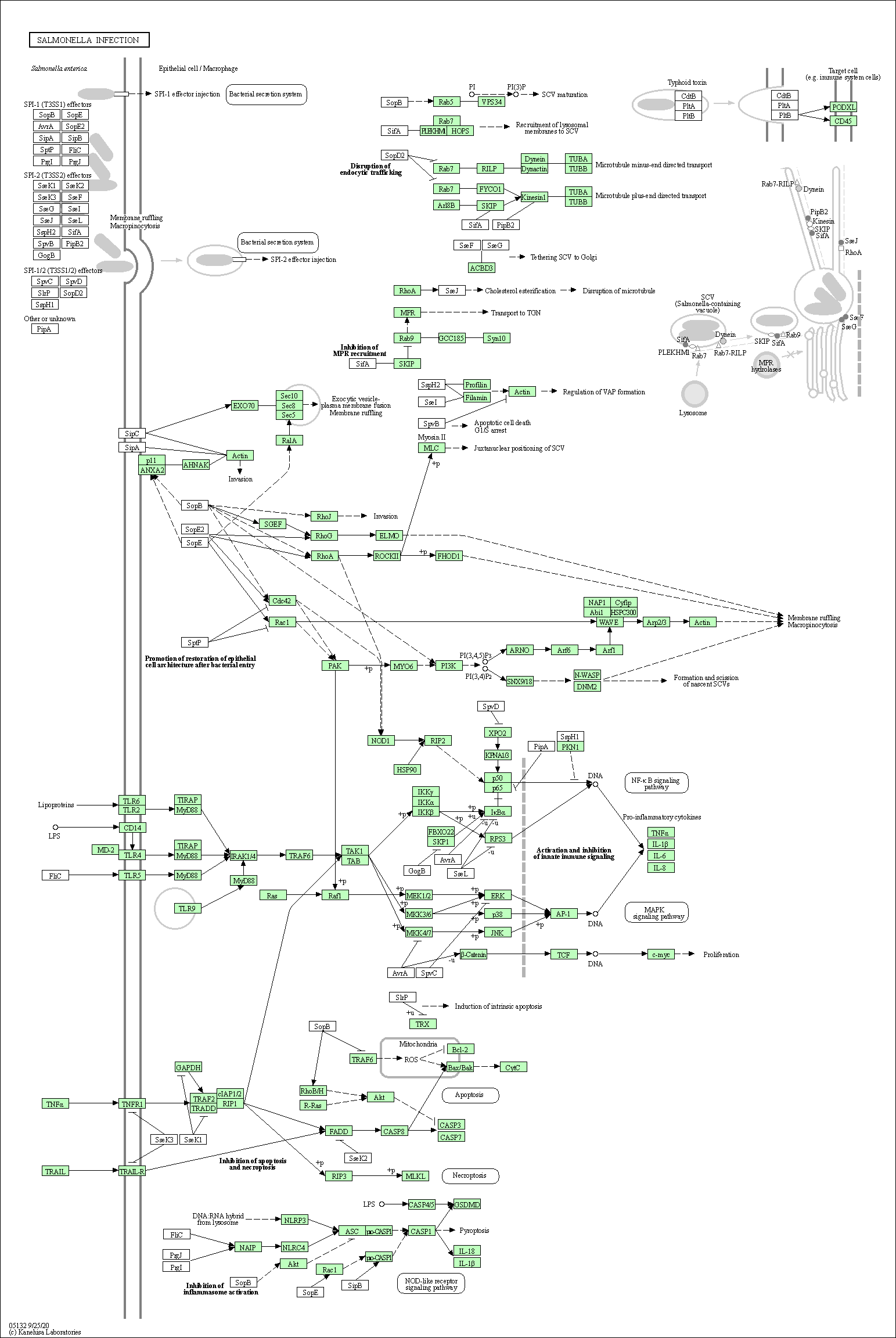
|
|||||||
| Yersinia infection | hsa05135 |
Pathway Map 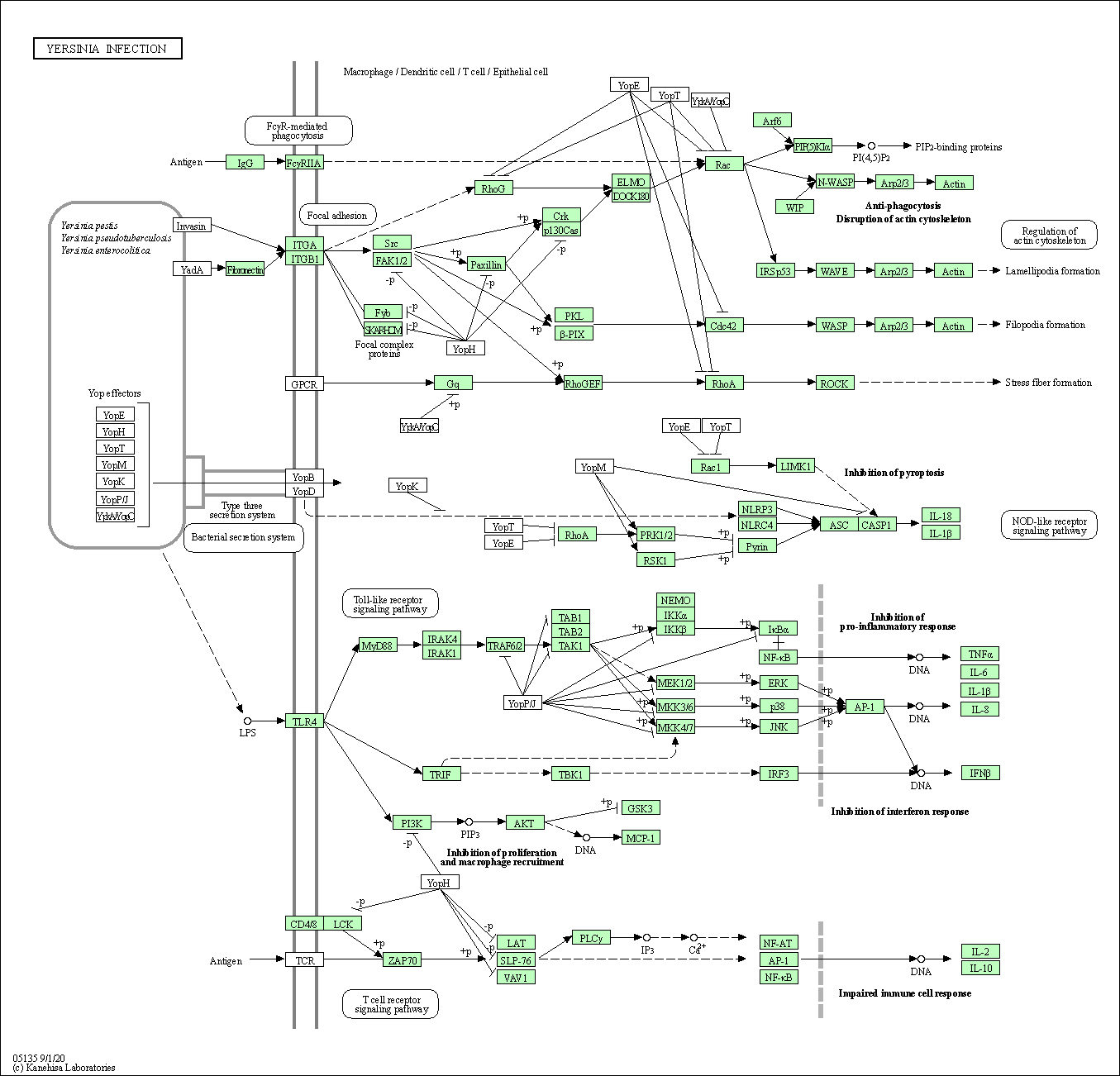
|
|||||||
| Hepatitis C | hsa05160 |
Pathway Map 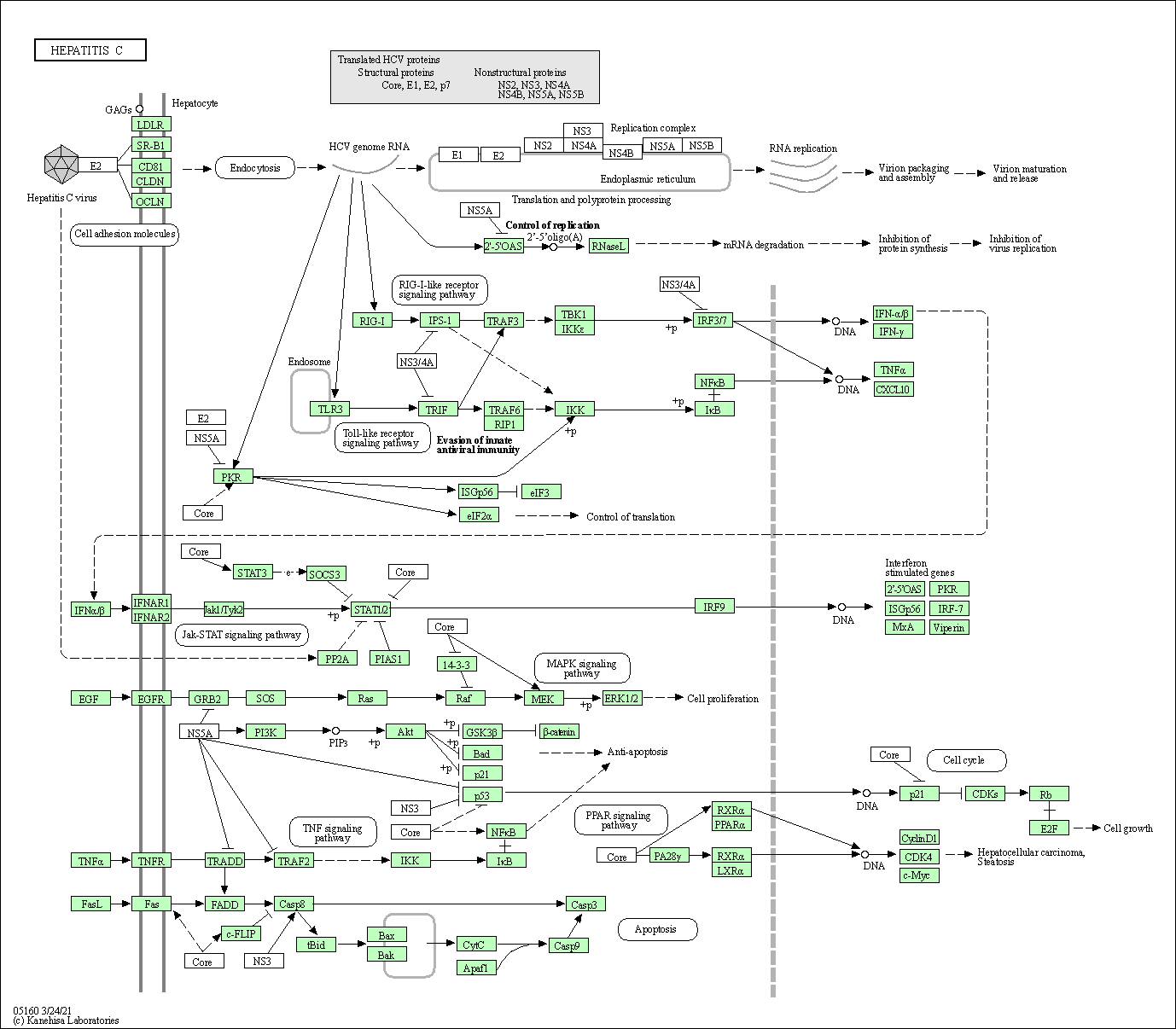
|
|||||||
| Hepatitis B | hsa05161 |
Pathway Map 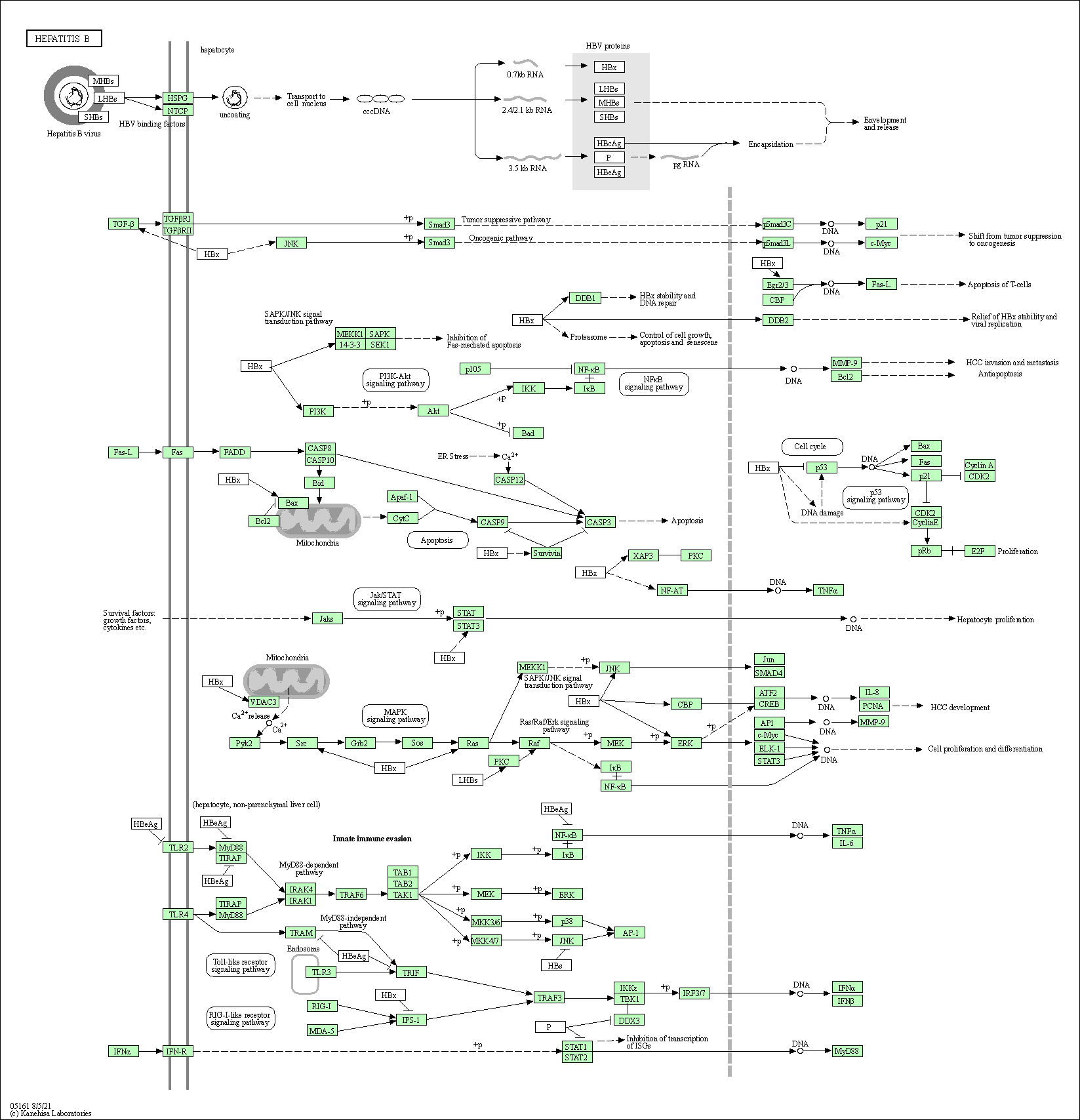
|
|||||||
| Human cytomegalovirus infection | hsa05163 |
Pathway Map 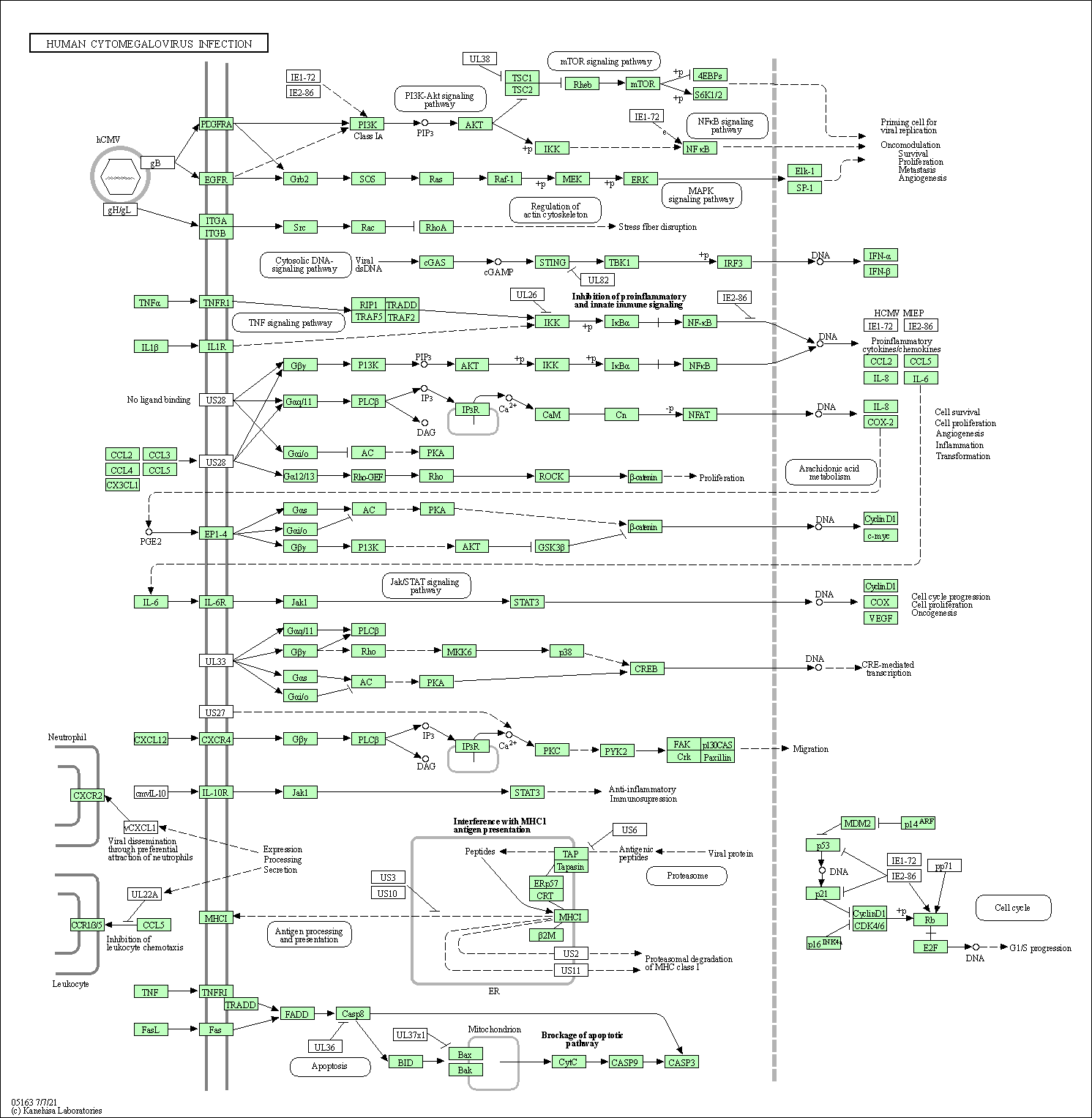
|
|||||||
| Influenza A | hsa05164 |
Pathway Map 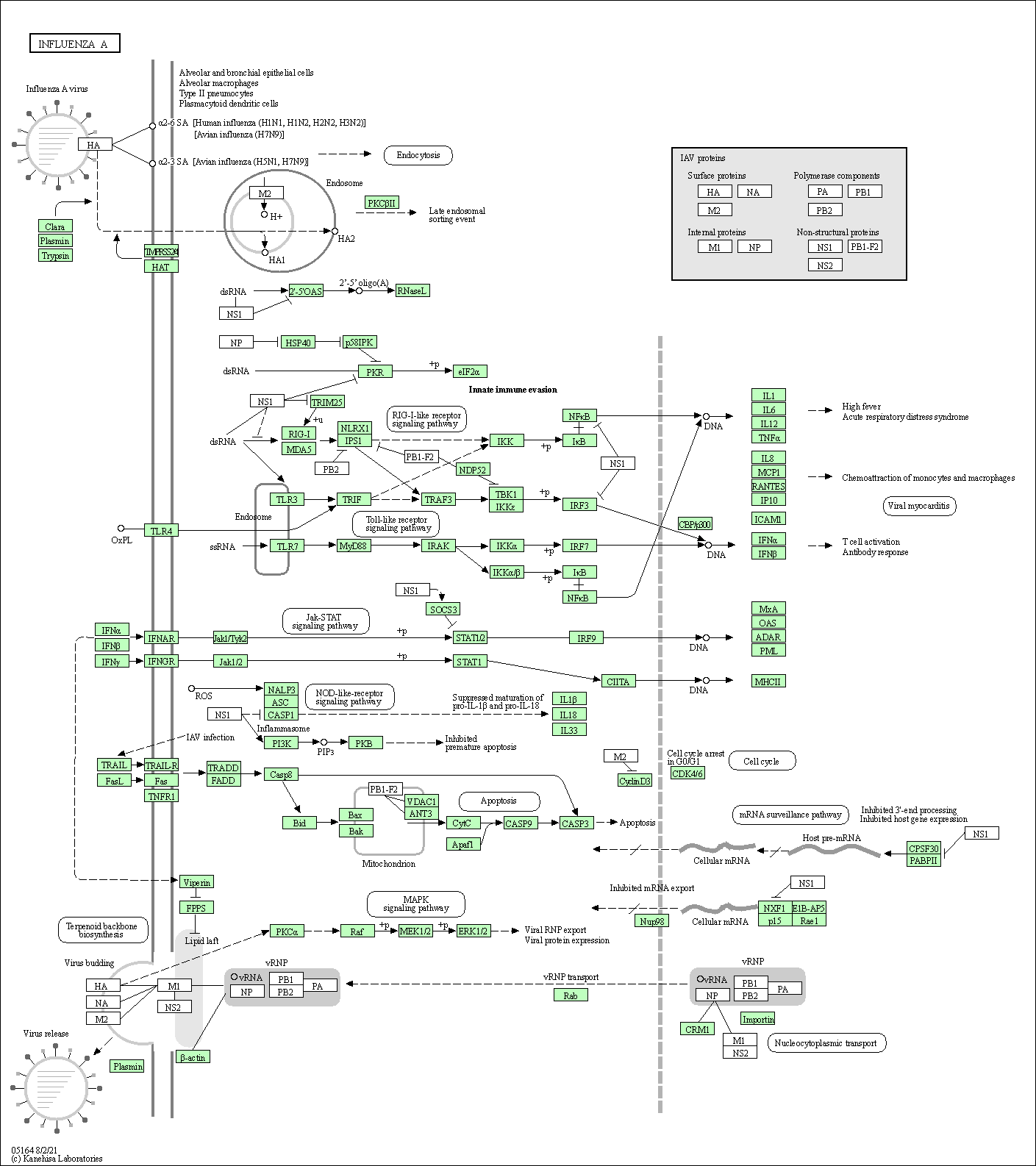
|
|||||||
| Human papillomavirus infection | hsa05165 |
Pathway Map 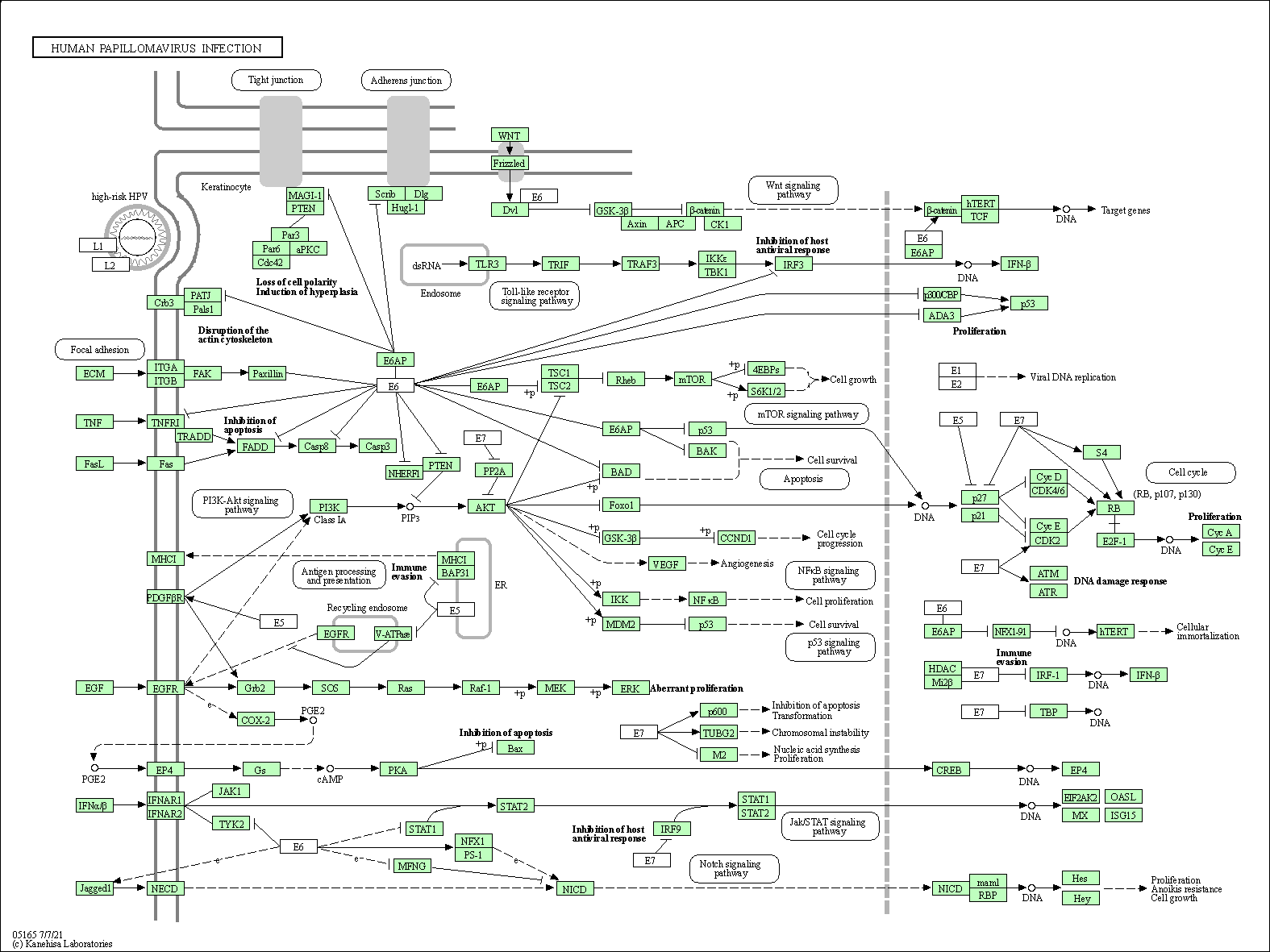
|
|||||||
| Human T-cell leukemia virus 1 infection | hsa05166 |
Pathway Map 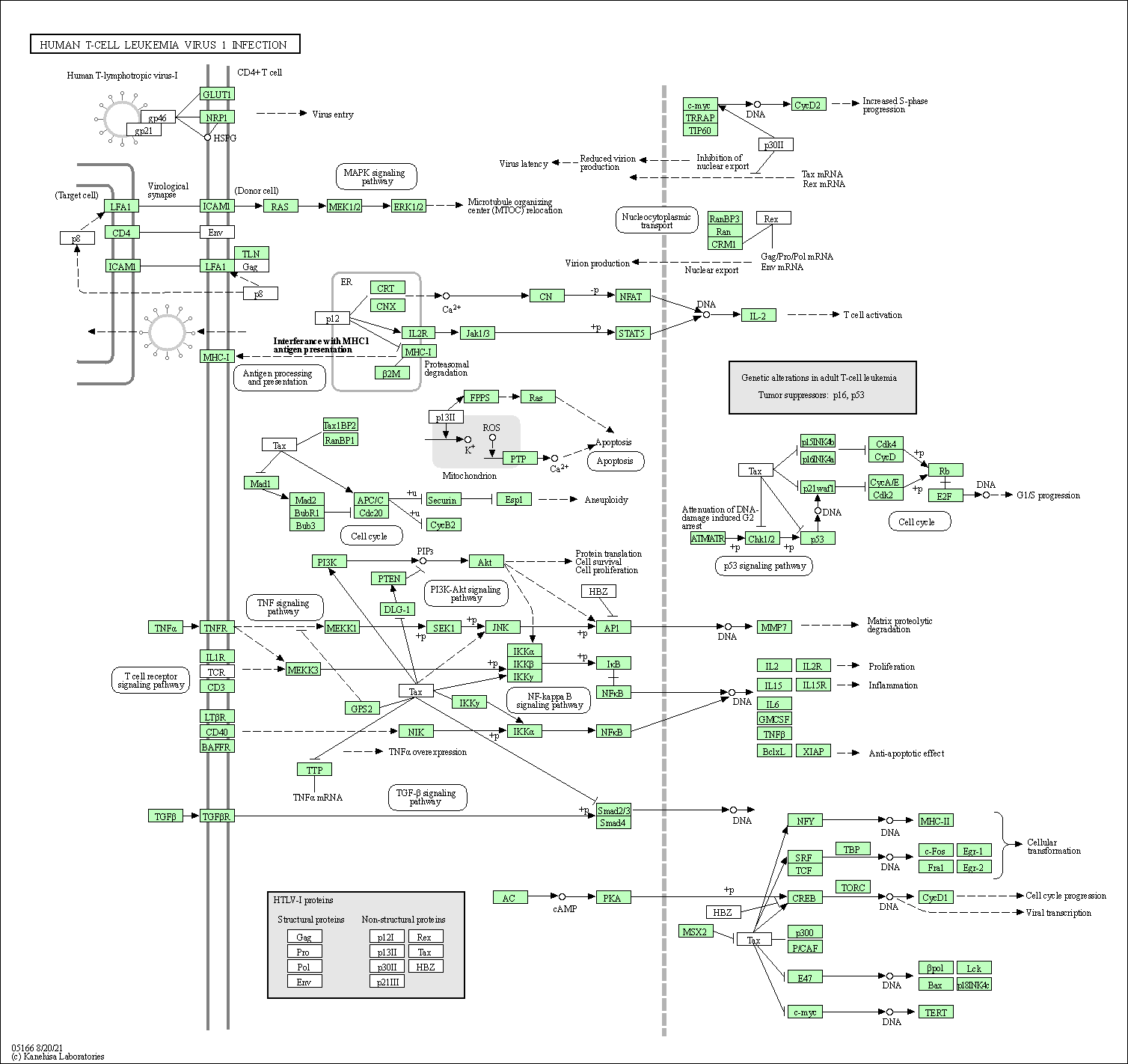
|
|||||||
| Kaposi sarcoma-associated herpesvirus infection | hsa05167 |
Pathway Map 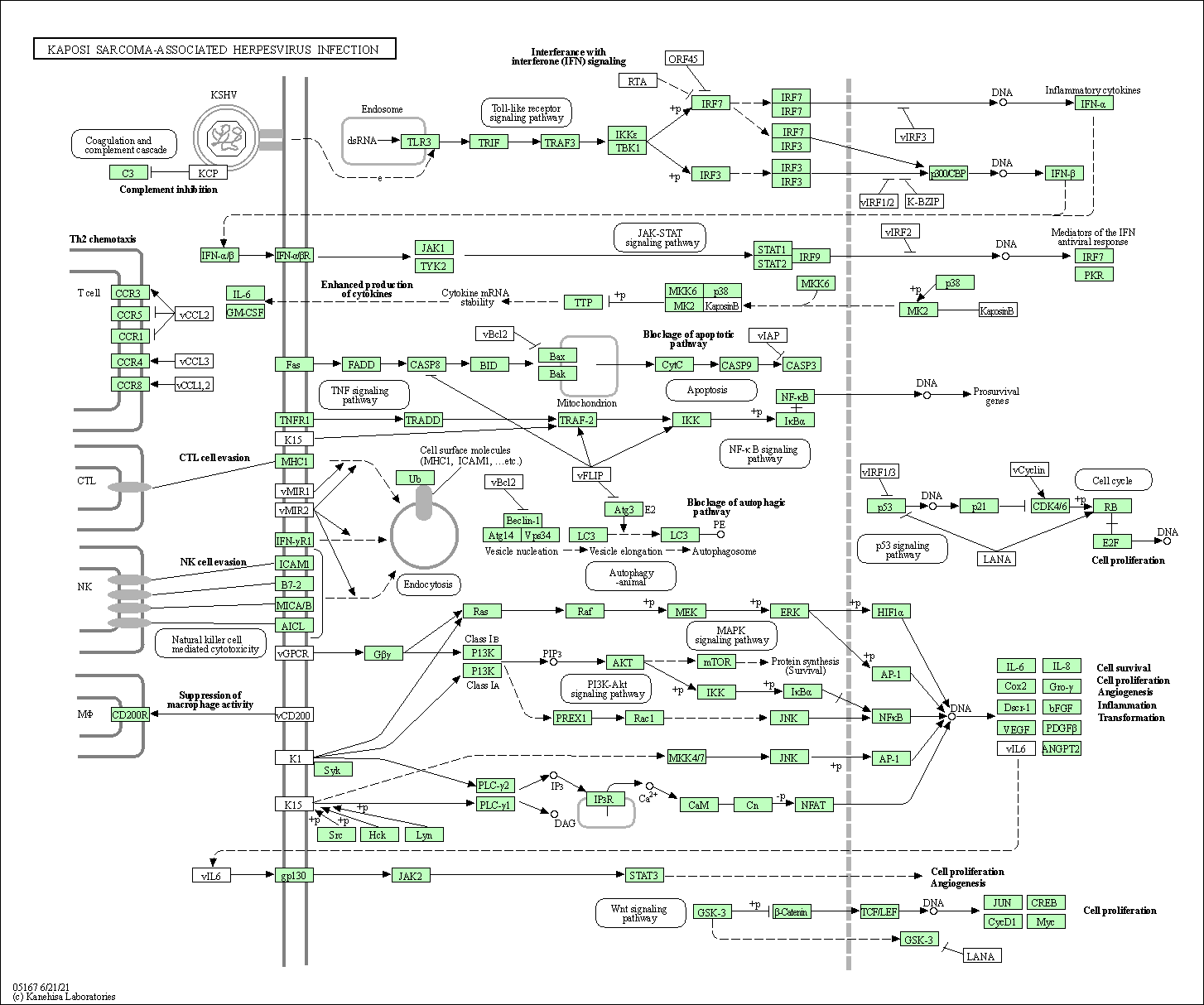
|
|||||||
| Human immunodeficiency virus 1 infection | hsa05170 |
Pathway Map 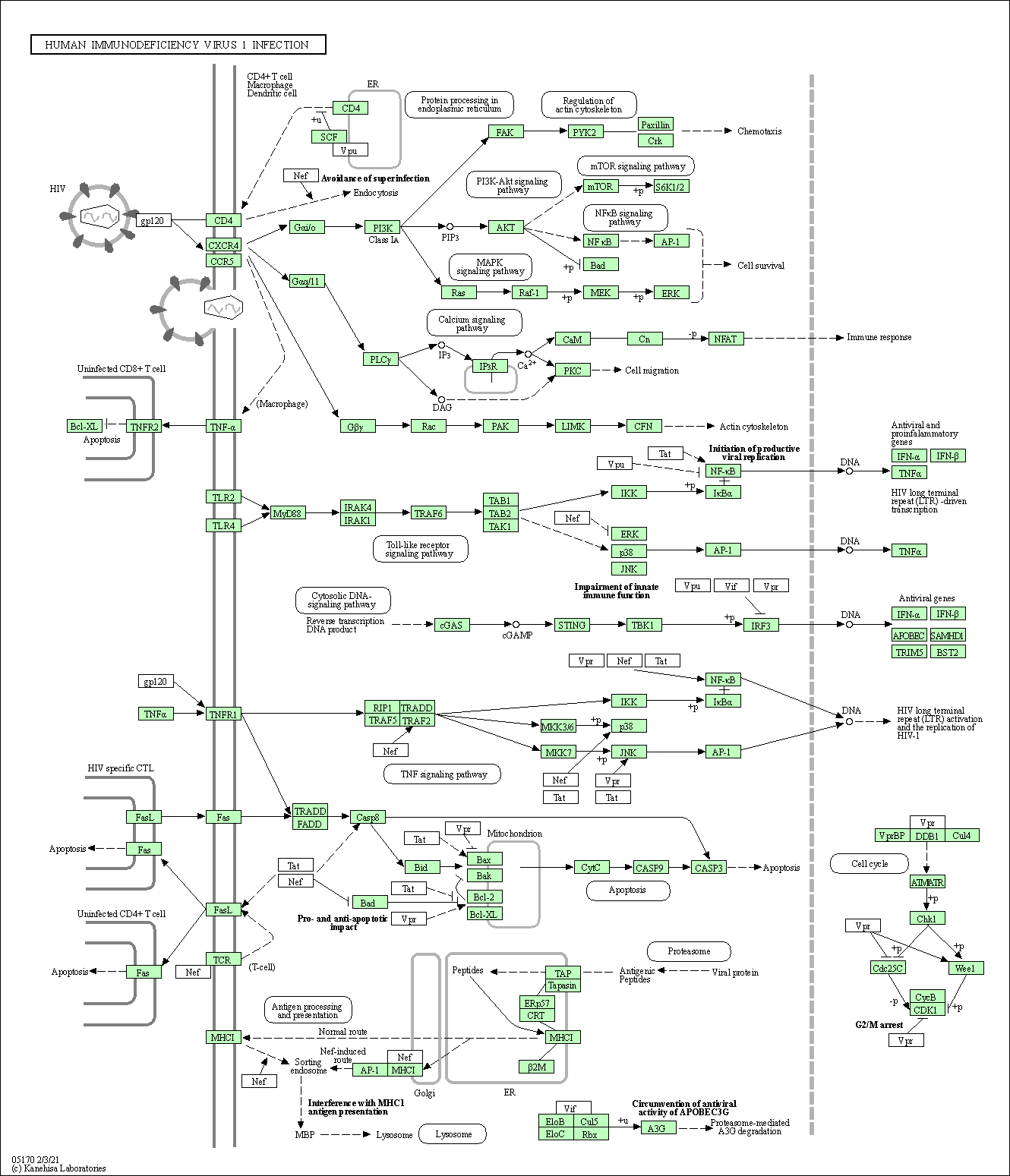
|
|||||||
| EGFR tyrosine kinase inhibitor resistance | hsa01521 |
Pathway Map 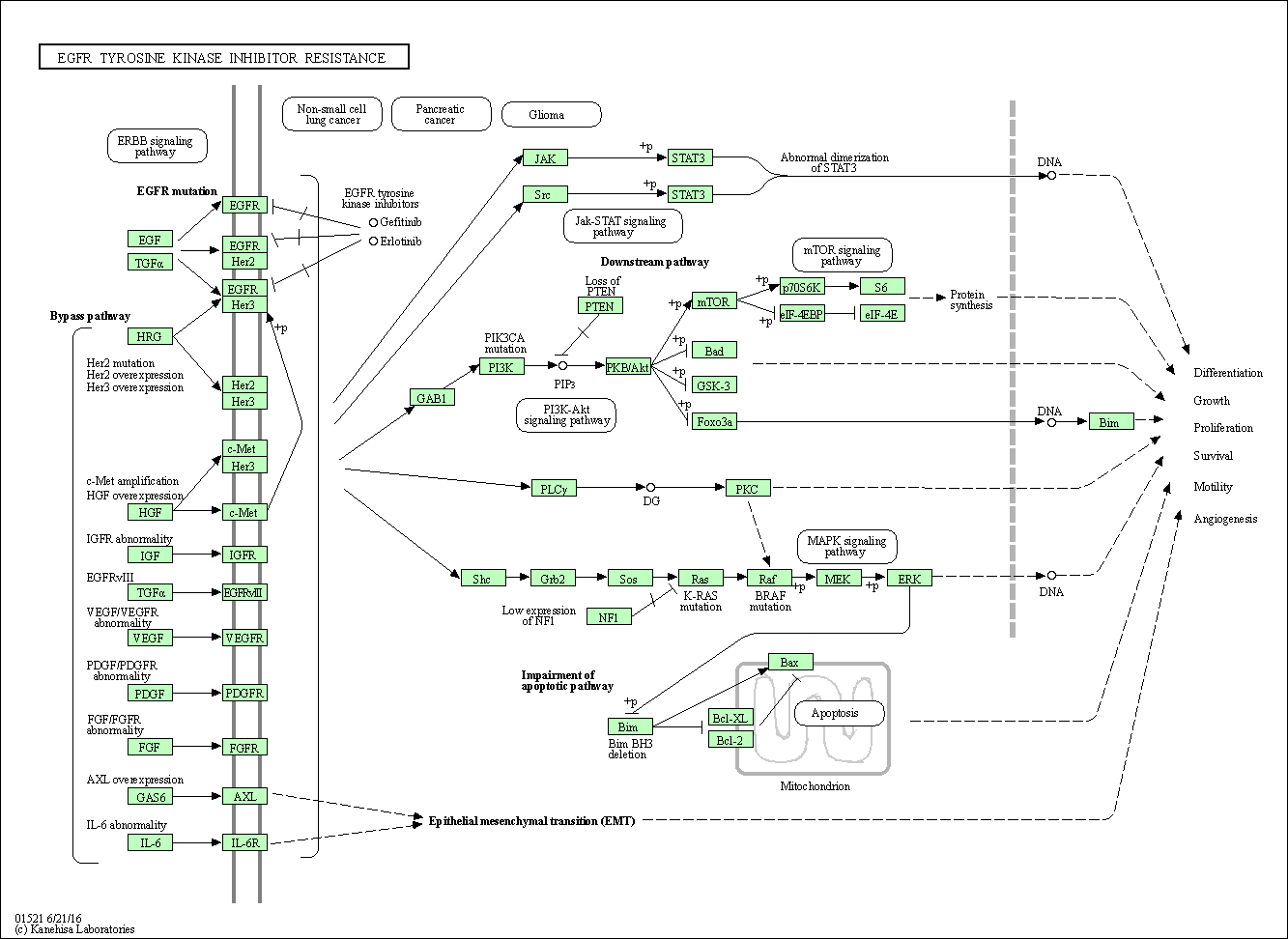
|
|||||||
| Endocrine resistance | hsa01522 |
Pathway Map 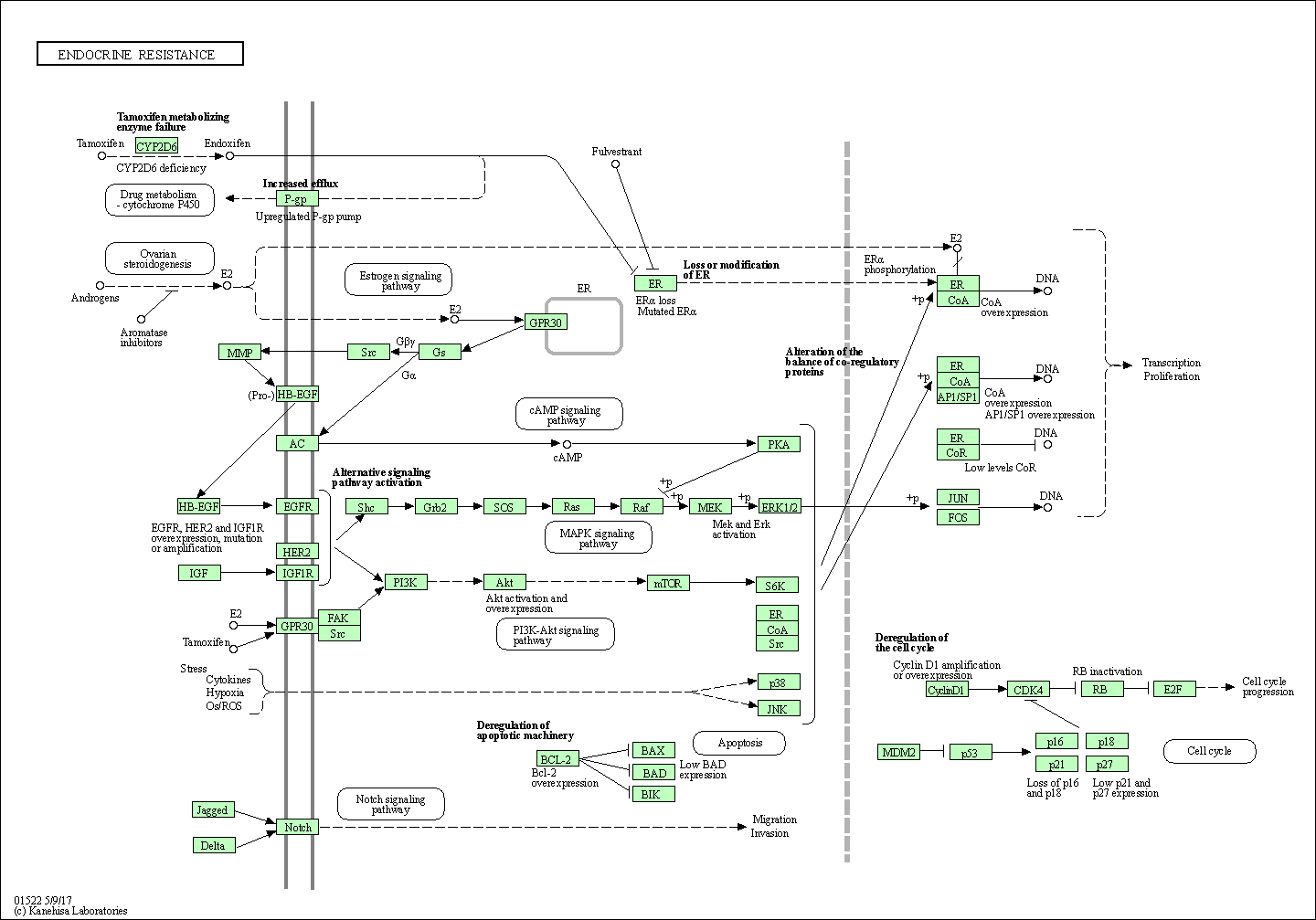
|
|||||||
| Pathways in cancer | hsa05200 |
Pathway Map 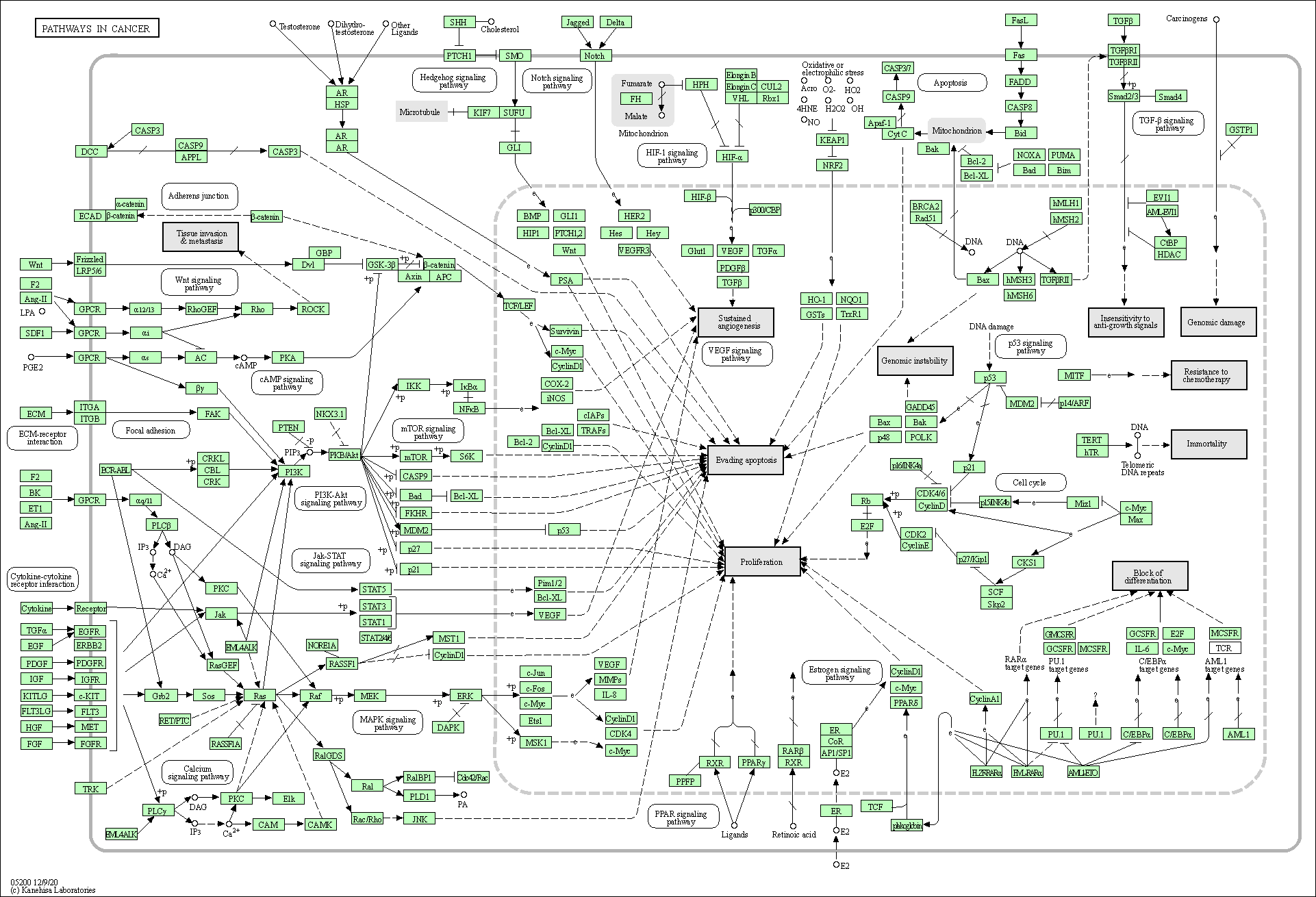
|
|||||||
| Proteoglycans in cancer | hsa05205 |
Pathway Map 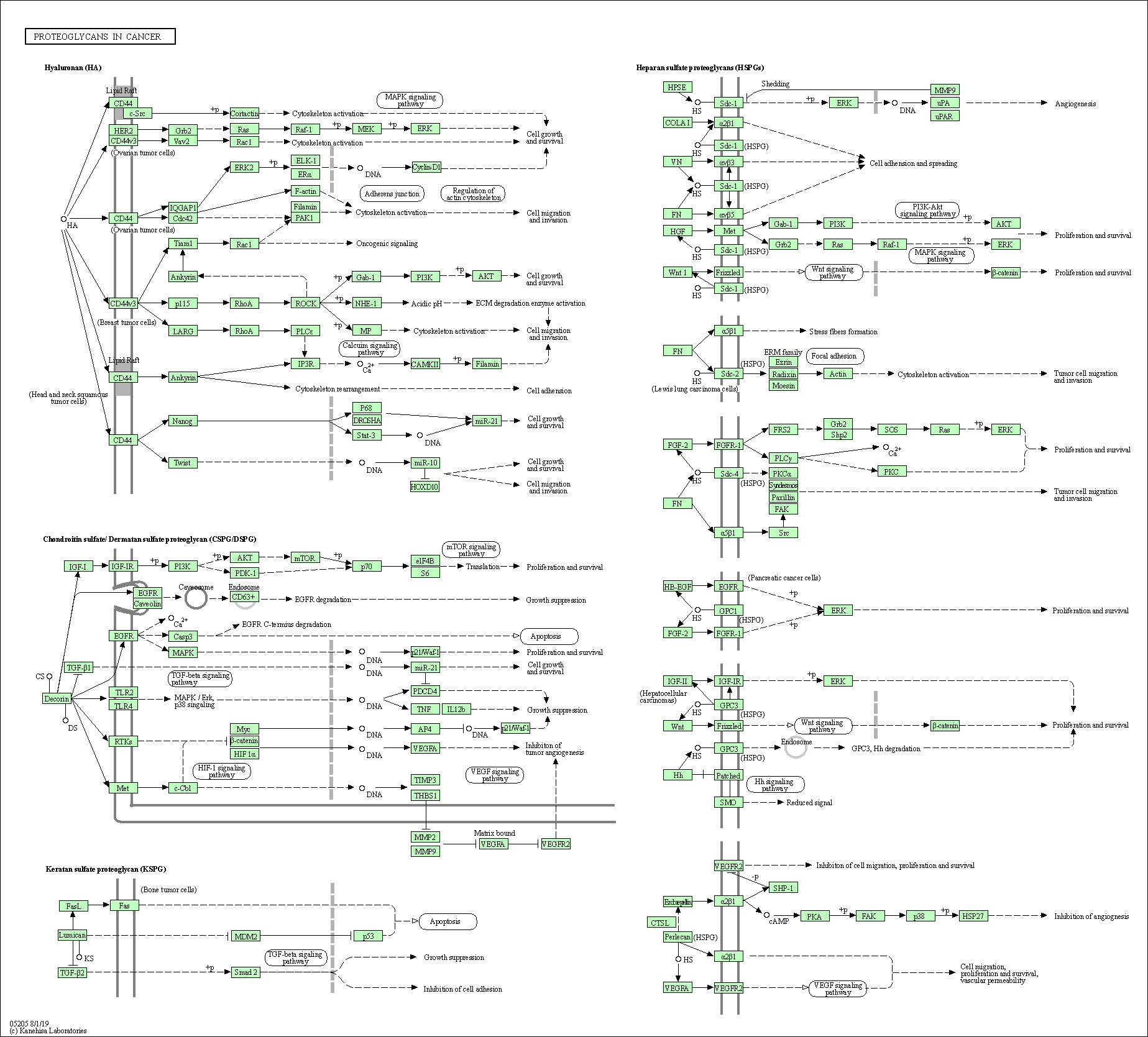
|
|||||||
| MicroRNAs in cancer | hsa05206 |
Pathway Map 
|
|||||||
| Chemical carcinogenesis - receptor activation | hsa05207 |
Pathway Map 
|
|||||||
| Chemical carcinogenesis - reactive oxygen species | hsa05208 |
Pathway Map 
|
|||||||
| Central carbon metabolism in cancer | hsa05230 |
Pathway Map 
|
|||||||
| Choline metabolism in cancer | hsa05231 |
Pathway Map 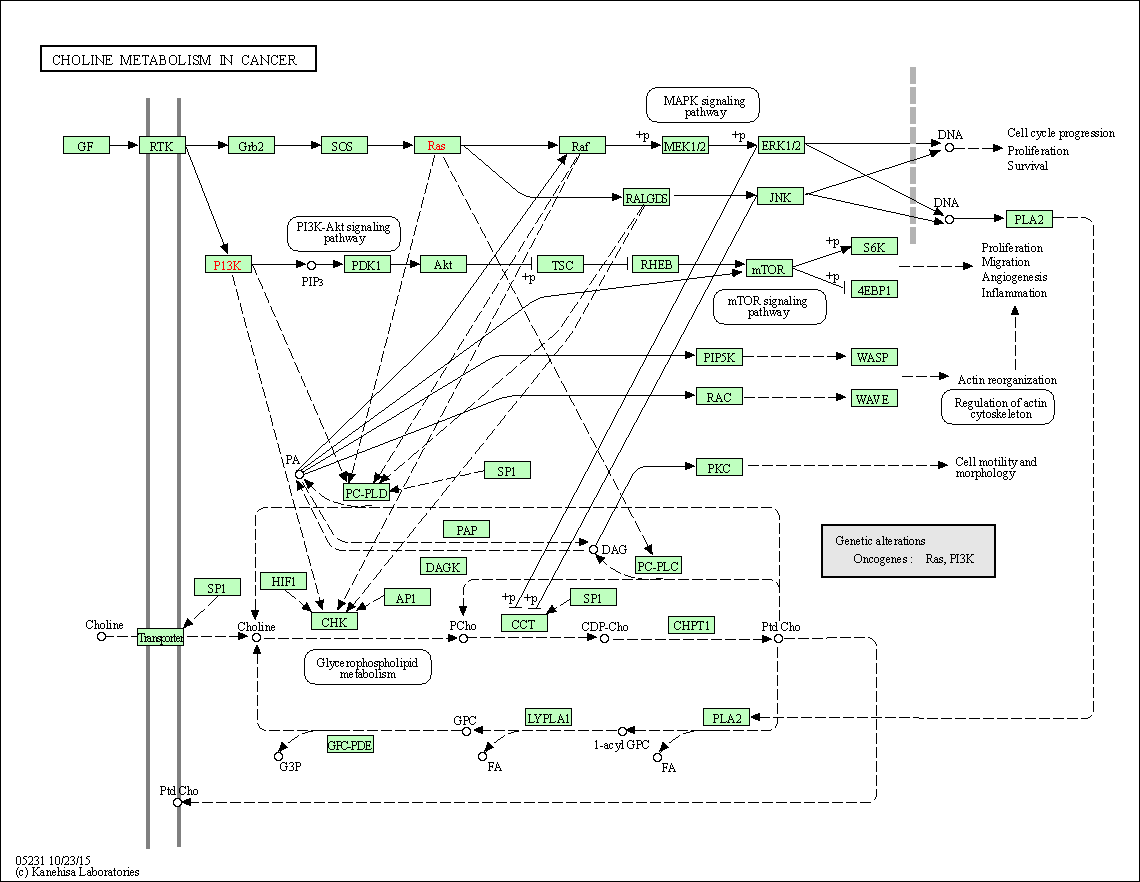
|
|||||||
| PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 |
Pathway Map 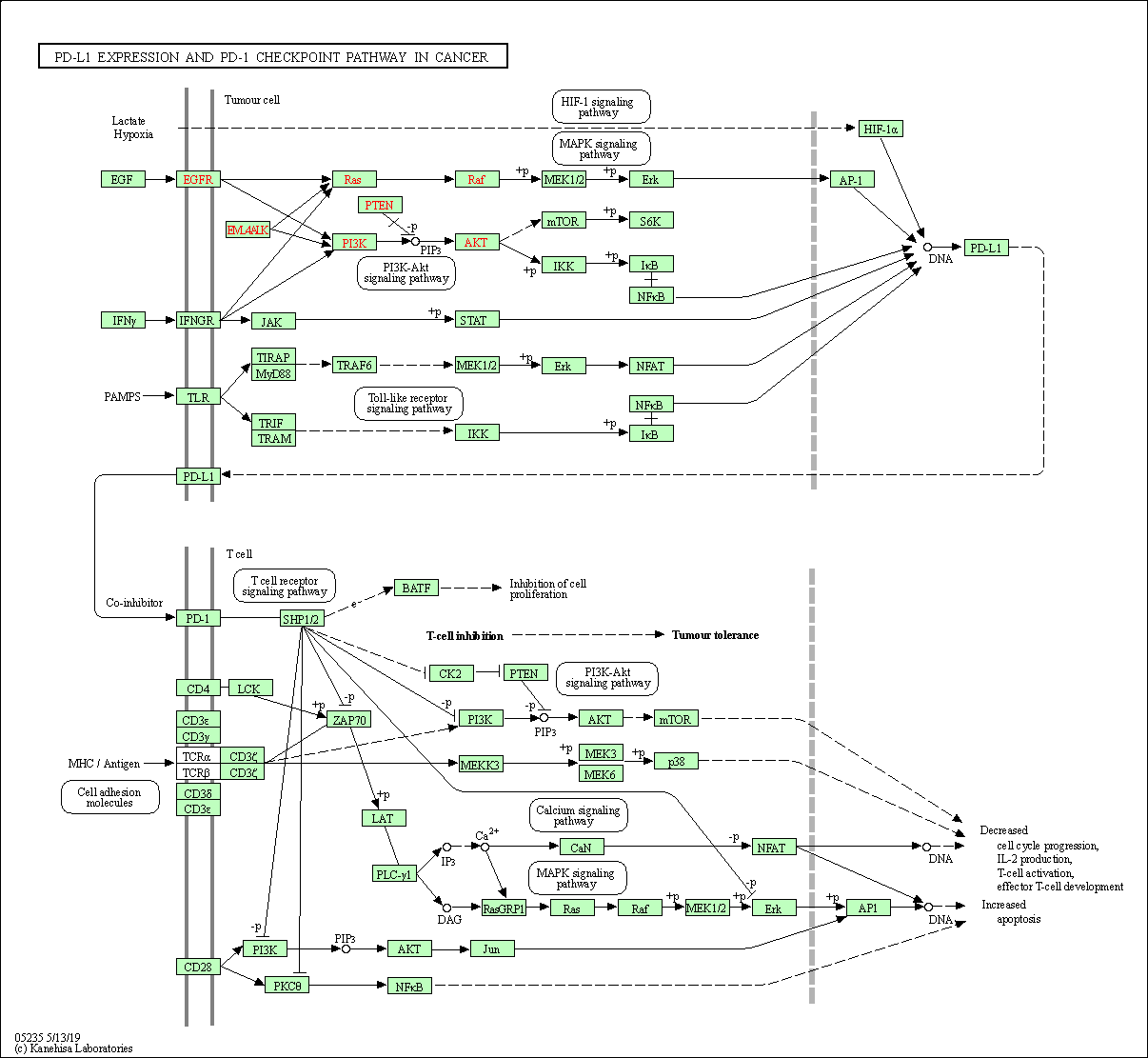
|
|||||||
| Colorectal cancer | hsa05210 |
Pathway Map 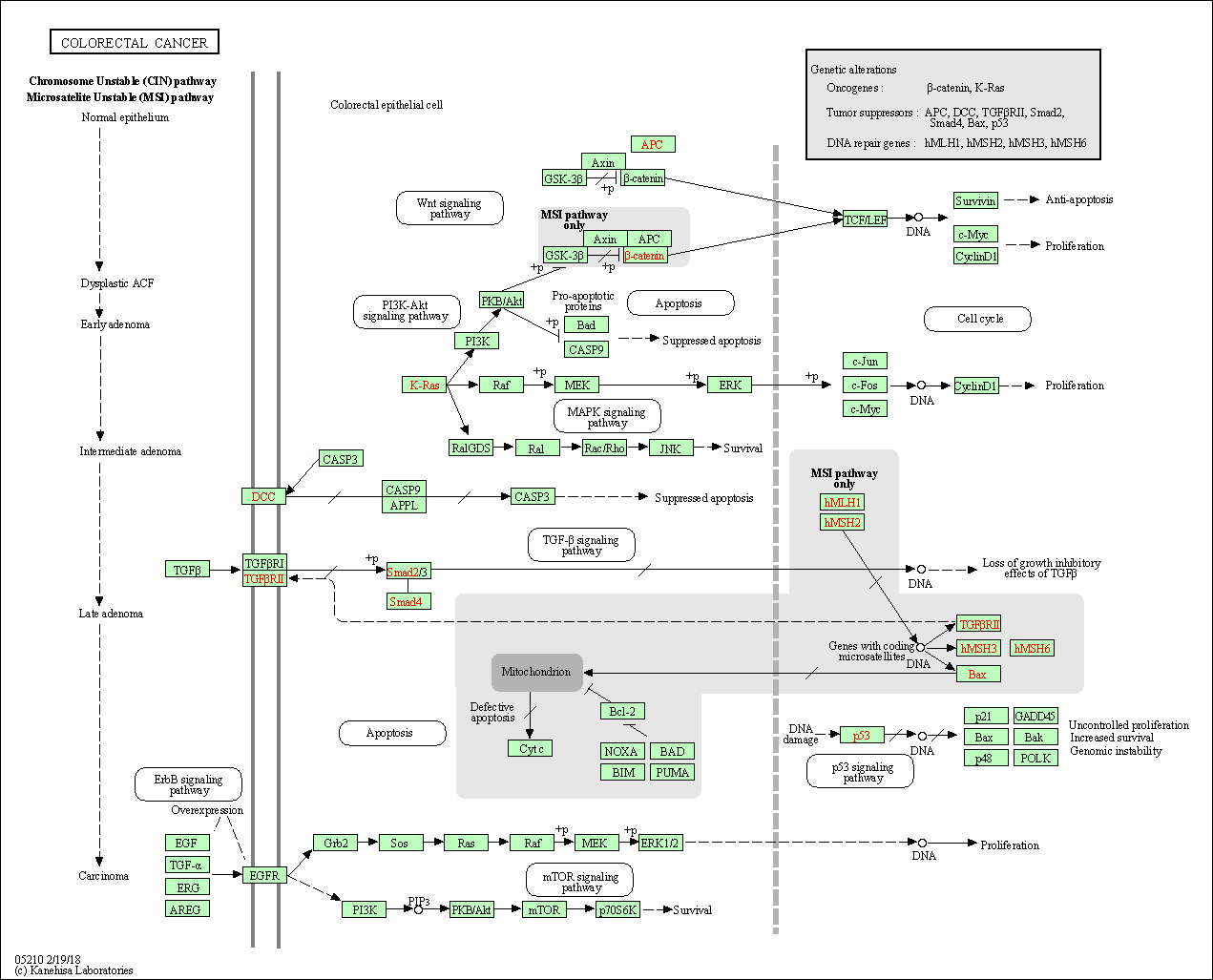
|
|||||||
| Renal cell carcinoma | hsa05211 |
Pathway Map 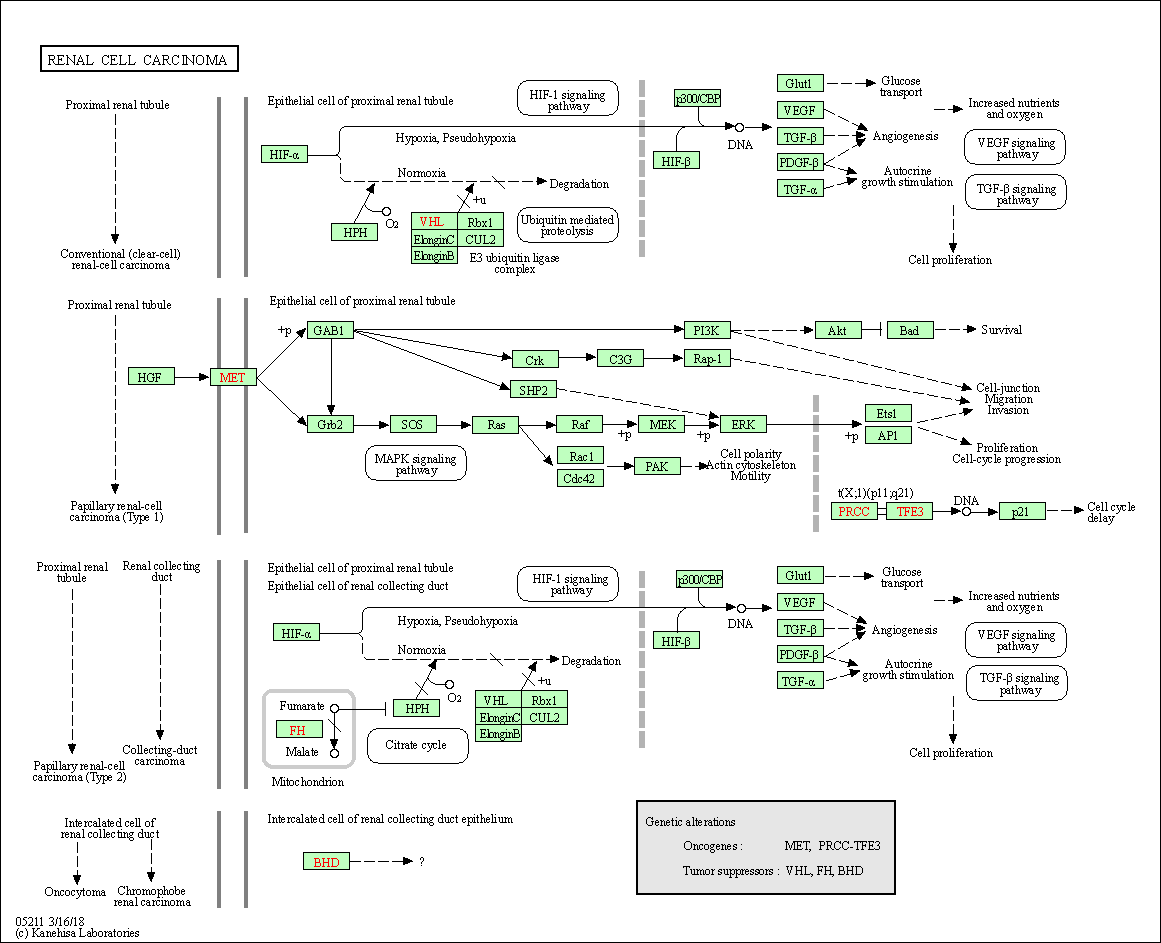
|
|||||||
| Endometrial cancer | hsa05213 |
Pathway Map 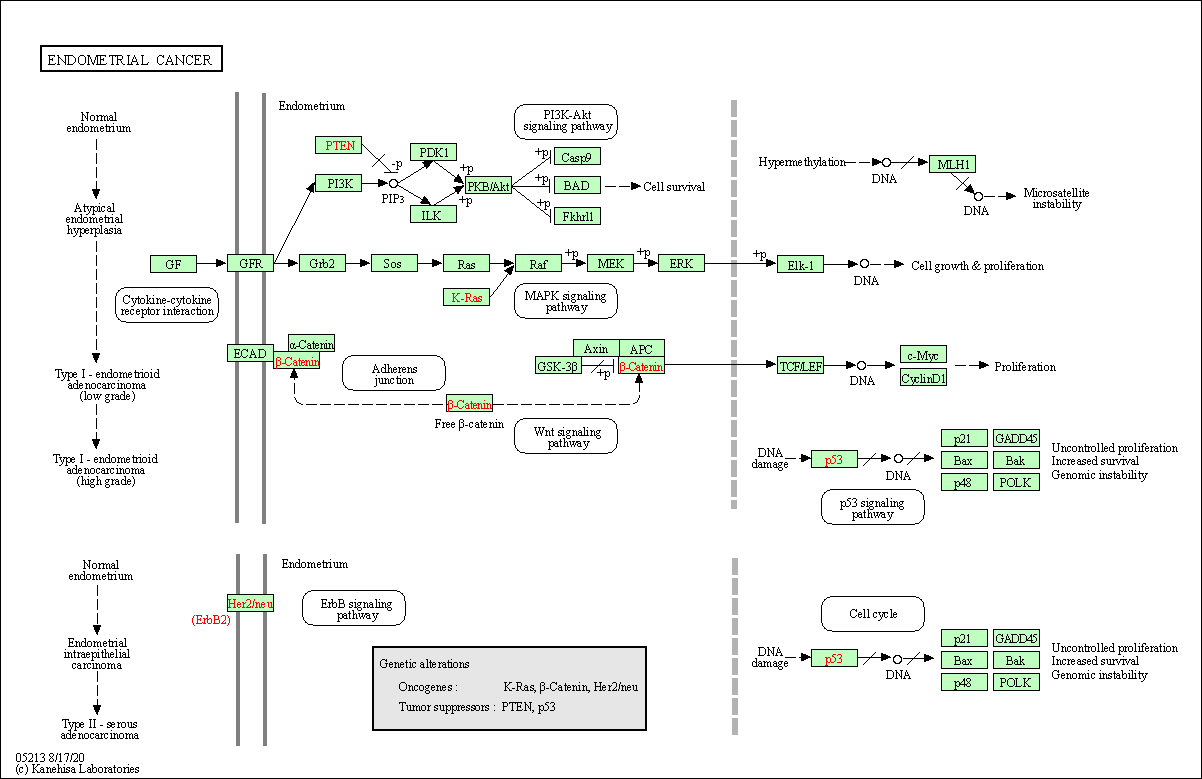
|
|||||||
| Glioma | hsa05214 |
Pathway Map 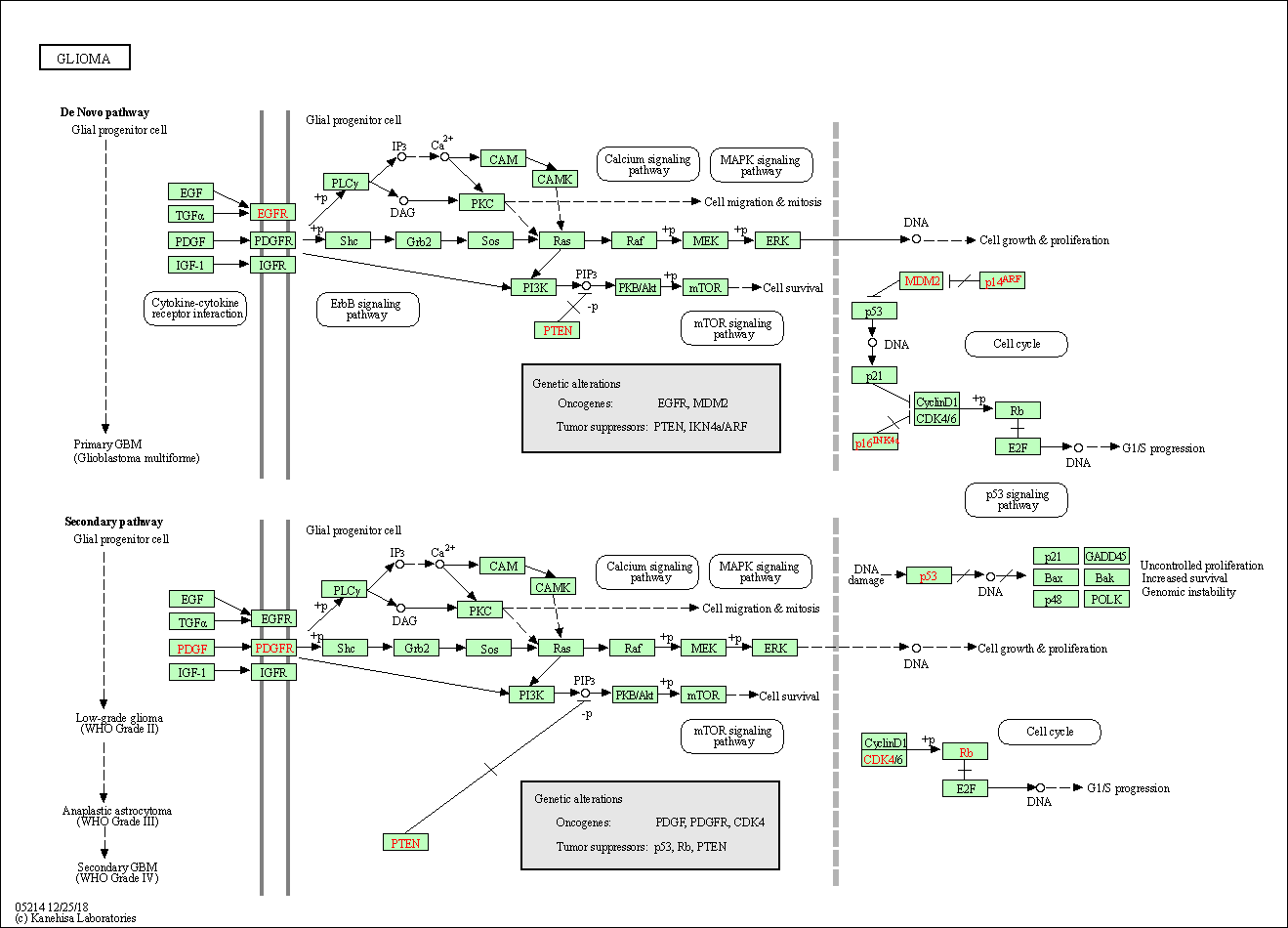
|
|||||||
| Prostate cancer | hsa05215 |
Pathway Map 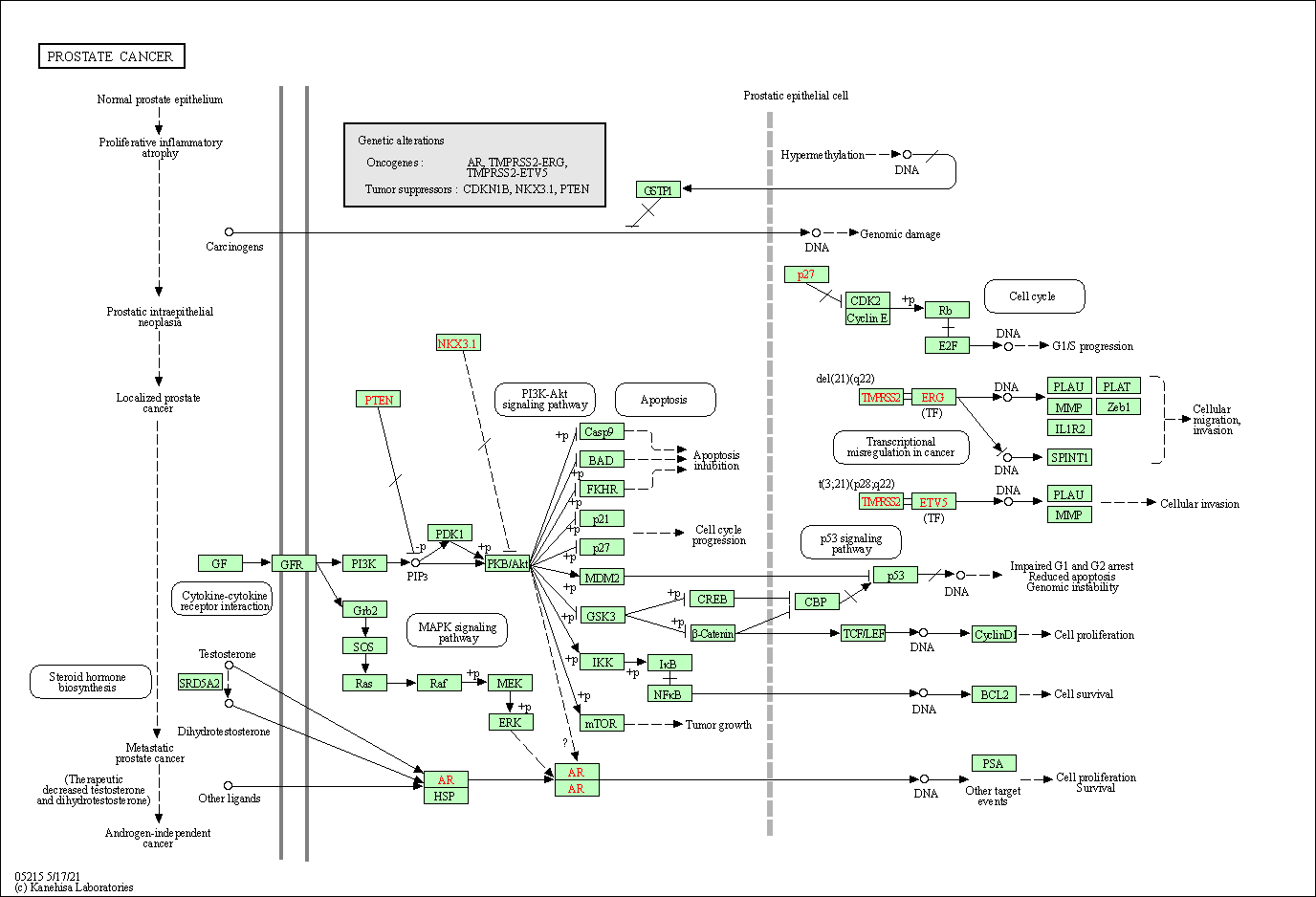
|
|||||||
| Thyroid cancer | hsa05216 |
Pathway Map 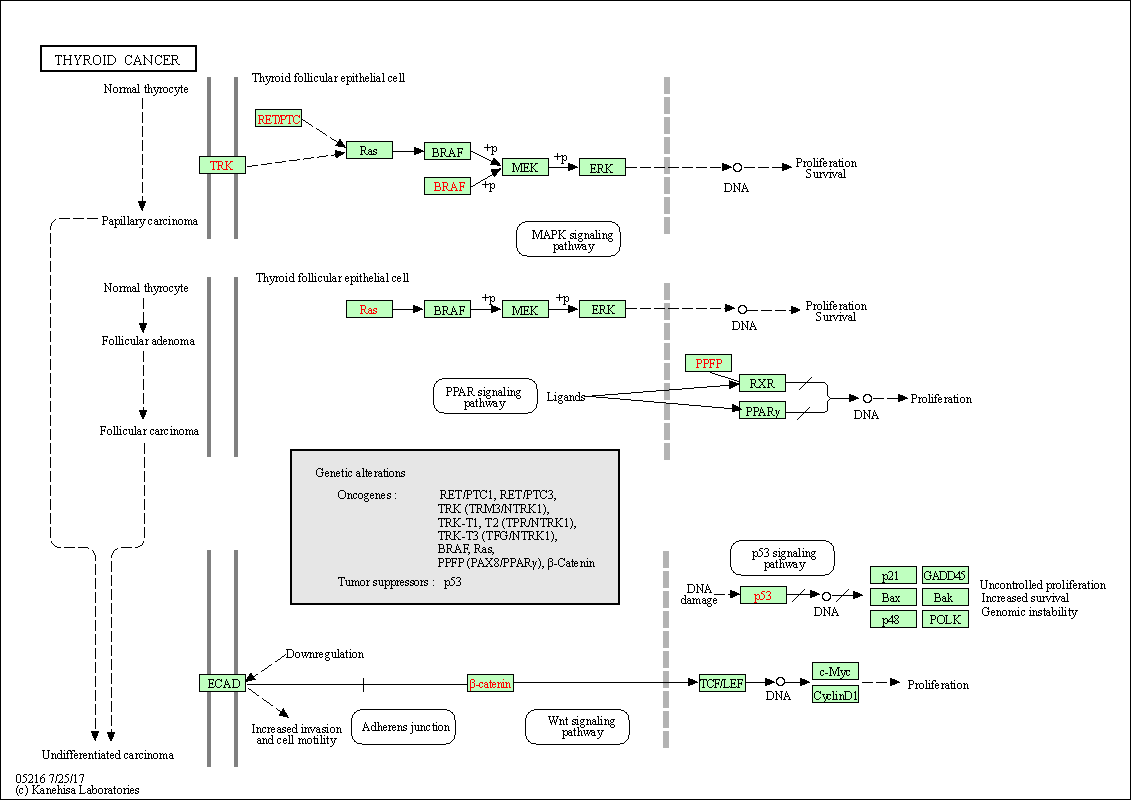
|
|||||||
| Melanoma | hsa05218 |
Pathway Map 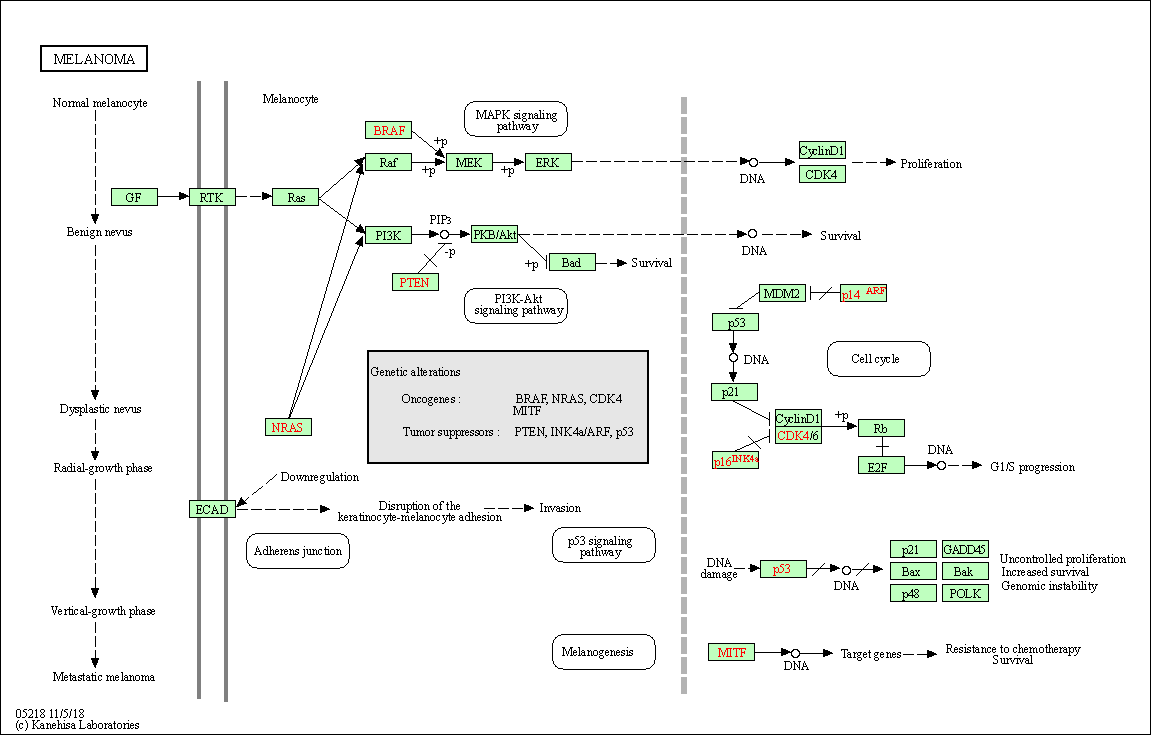
|
|||||||
| Bladder cancer | hsa05219 |
Pathway Map 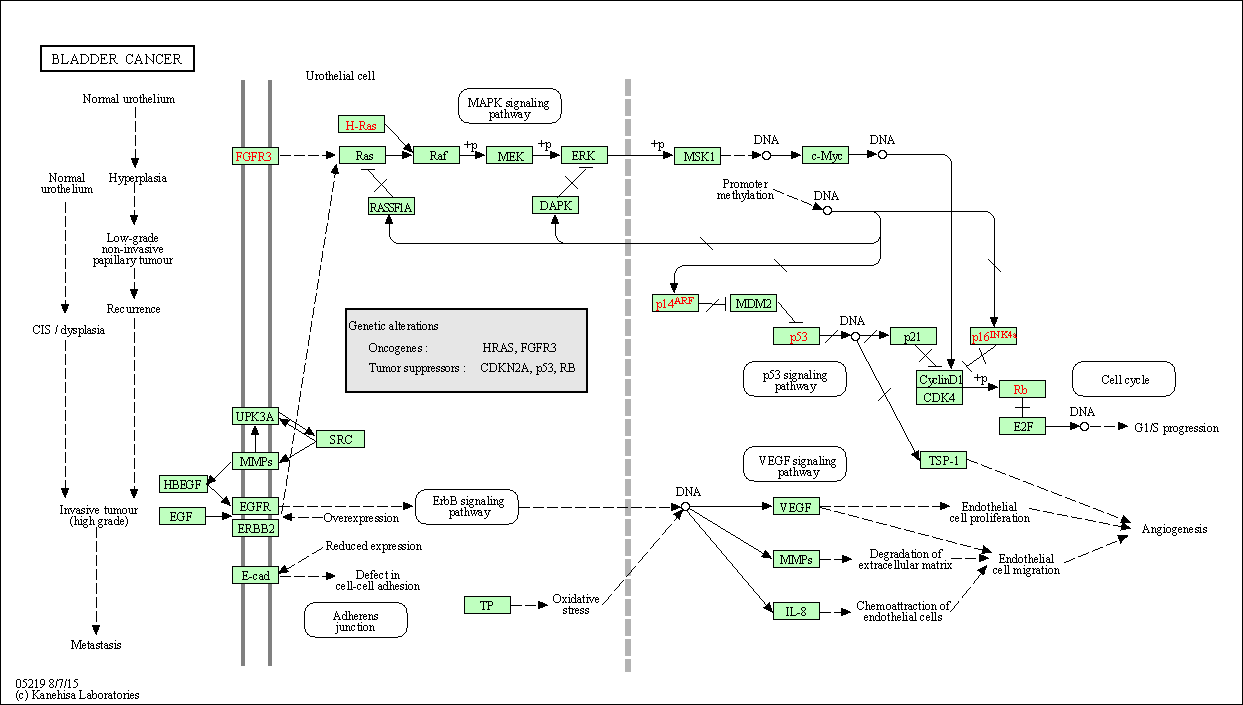
|
|||||||
| Chronic myeloid leukemia | hsa05220 |
Pathway Map 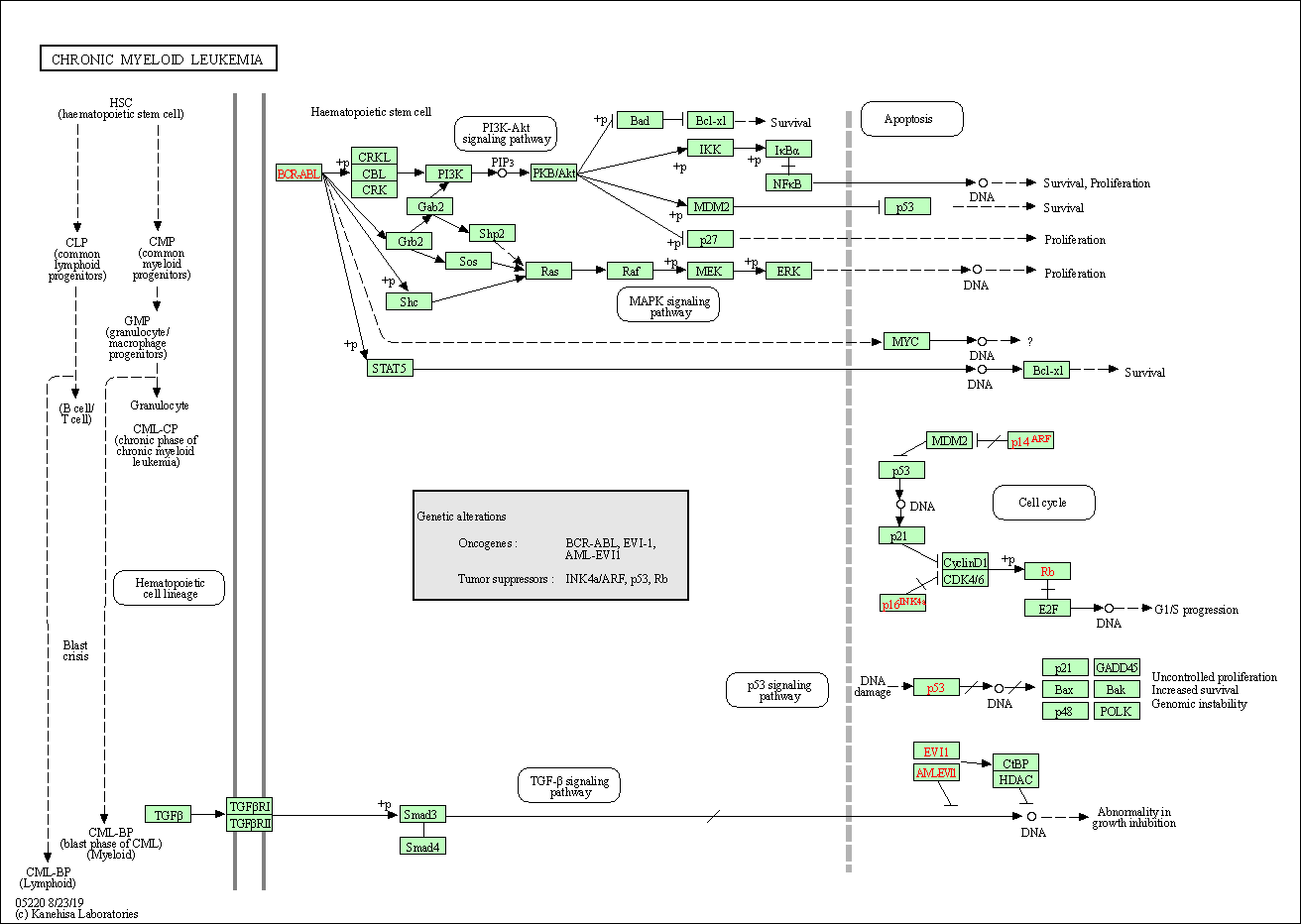
|
|||||||
| Acute myeloid leukemia | hsa05221 |
Pathway Map 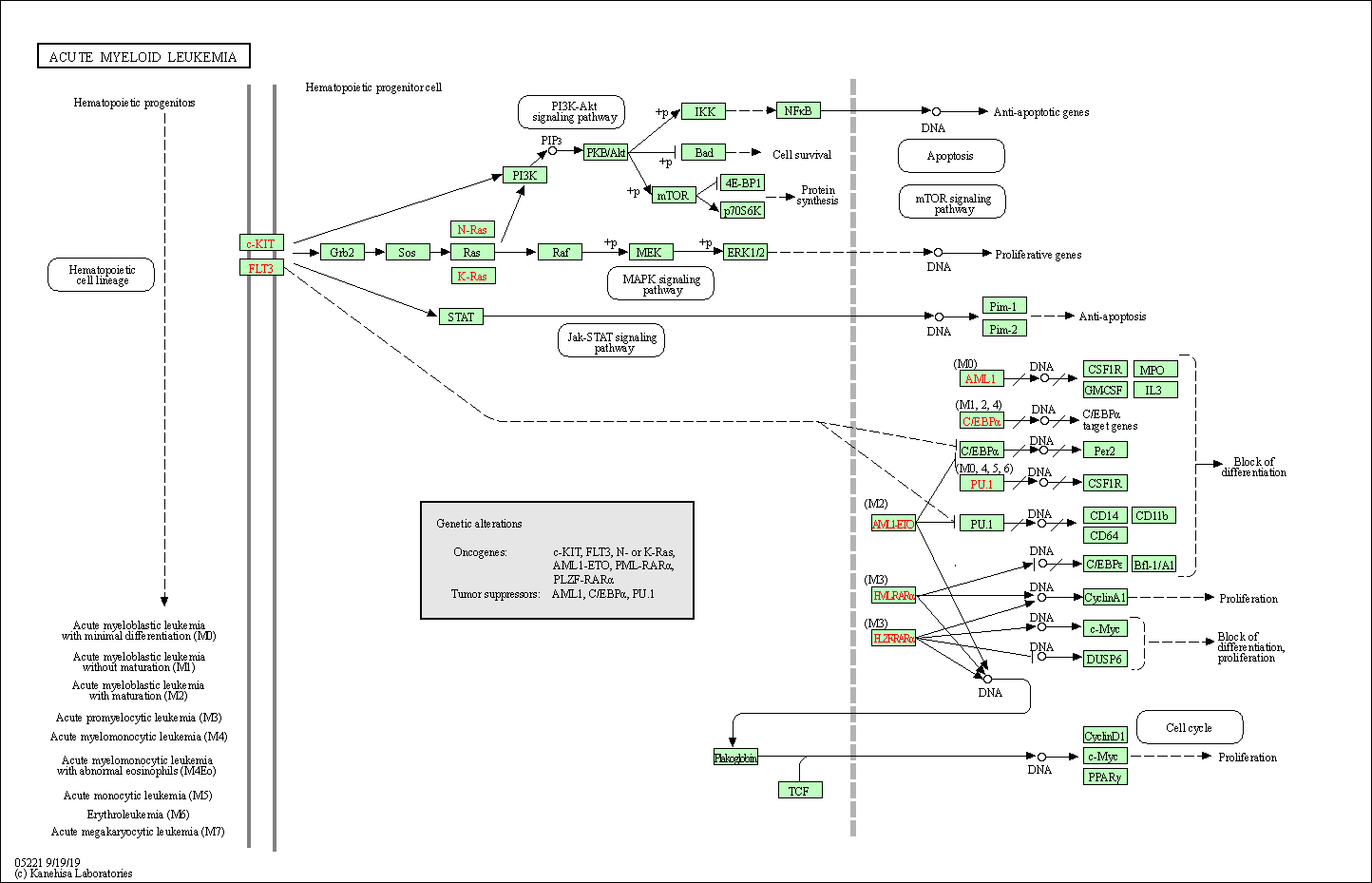
|
|||||||
| Non-small cell lung cancer | hsa05223 |
Pathway Map 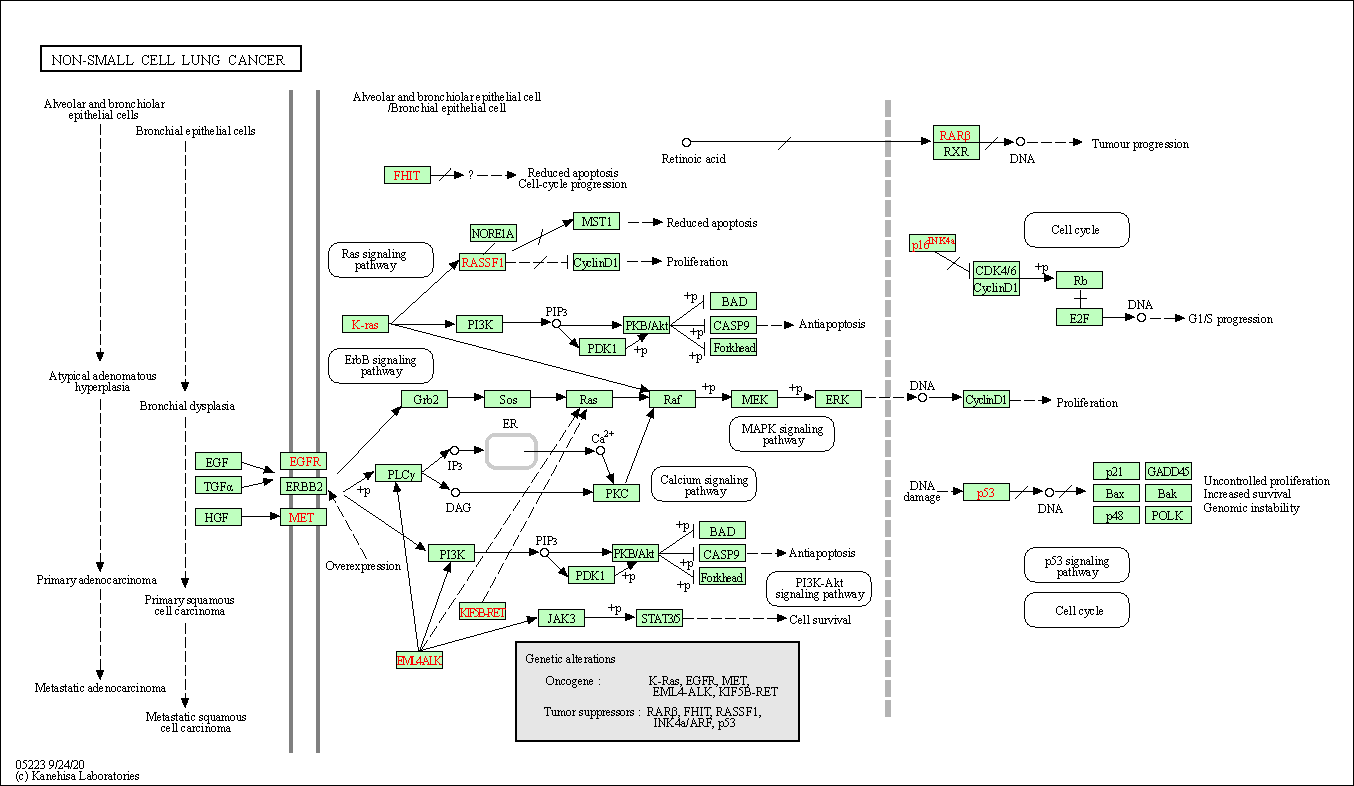
|
|||||||
| Breast cancer | hsa05224 |
Pathway Map 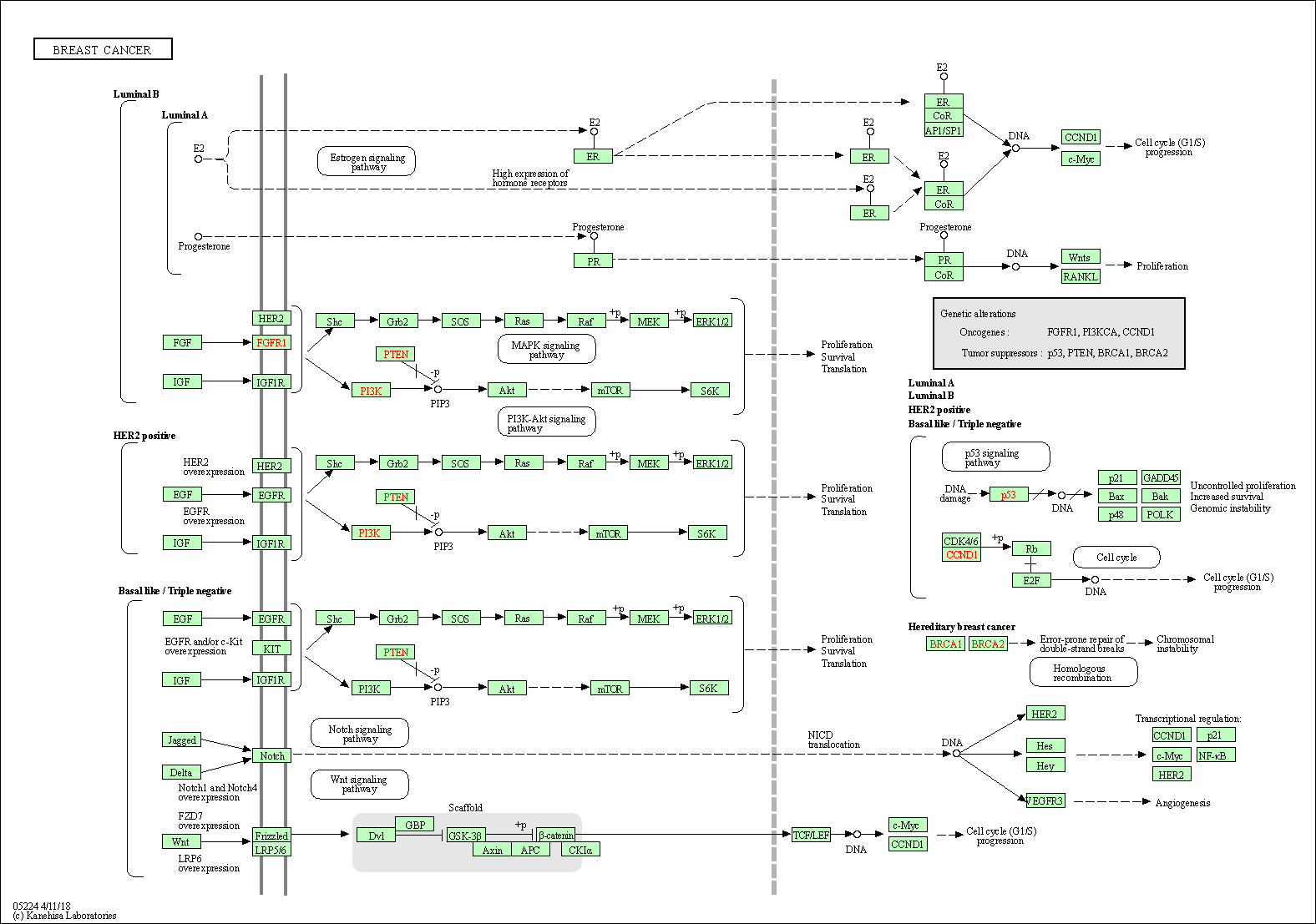
|
|||||||
| Hepatocellular carcinoma | hsa05225 |
Pathway Map 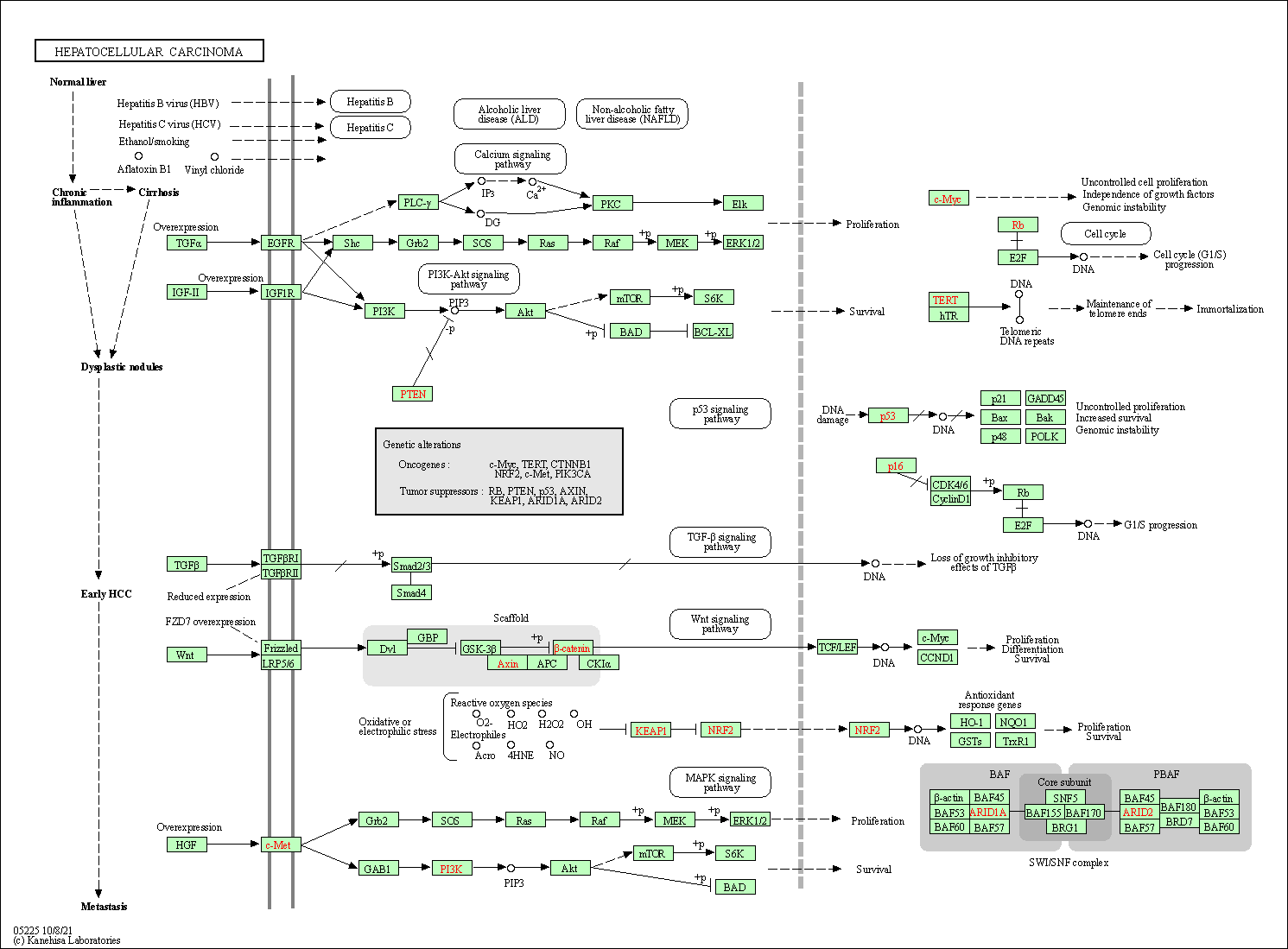
|
|||||||
| Gastric cancer | hsa05226 |
Pathway Map 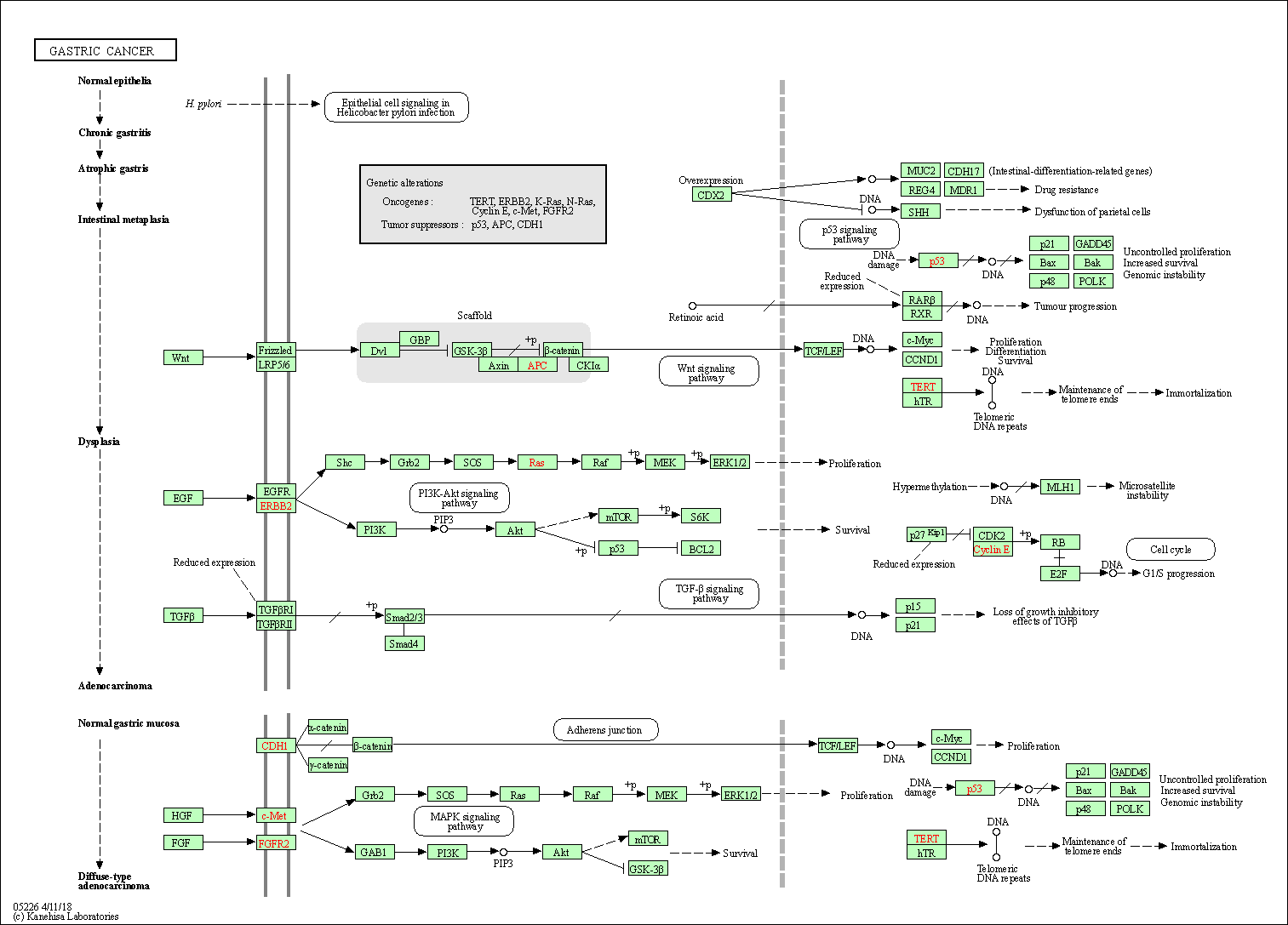
|
|||||||
| Apoptosis | hsa04210 |
Pathway Map 
|
|||||||
| Cellular senescence | hsa04218 |
Pathway Map 
|
|||||||
| Regulation of actin cytoskeleton | hsa04810 |
Pathway Map 
|
|||||||
| Gap junction | hsa04540 |
Pathway Map 
|
|||||||
| Signaling pathways regulating pluripotency of stem cells | hsa04550 |
Pathway Map 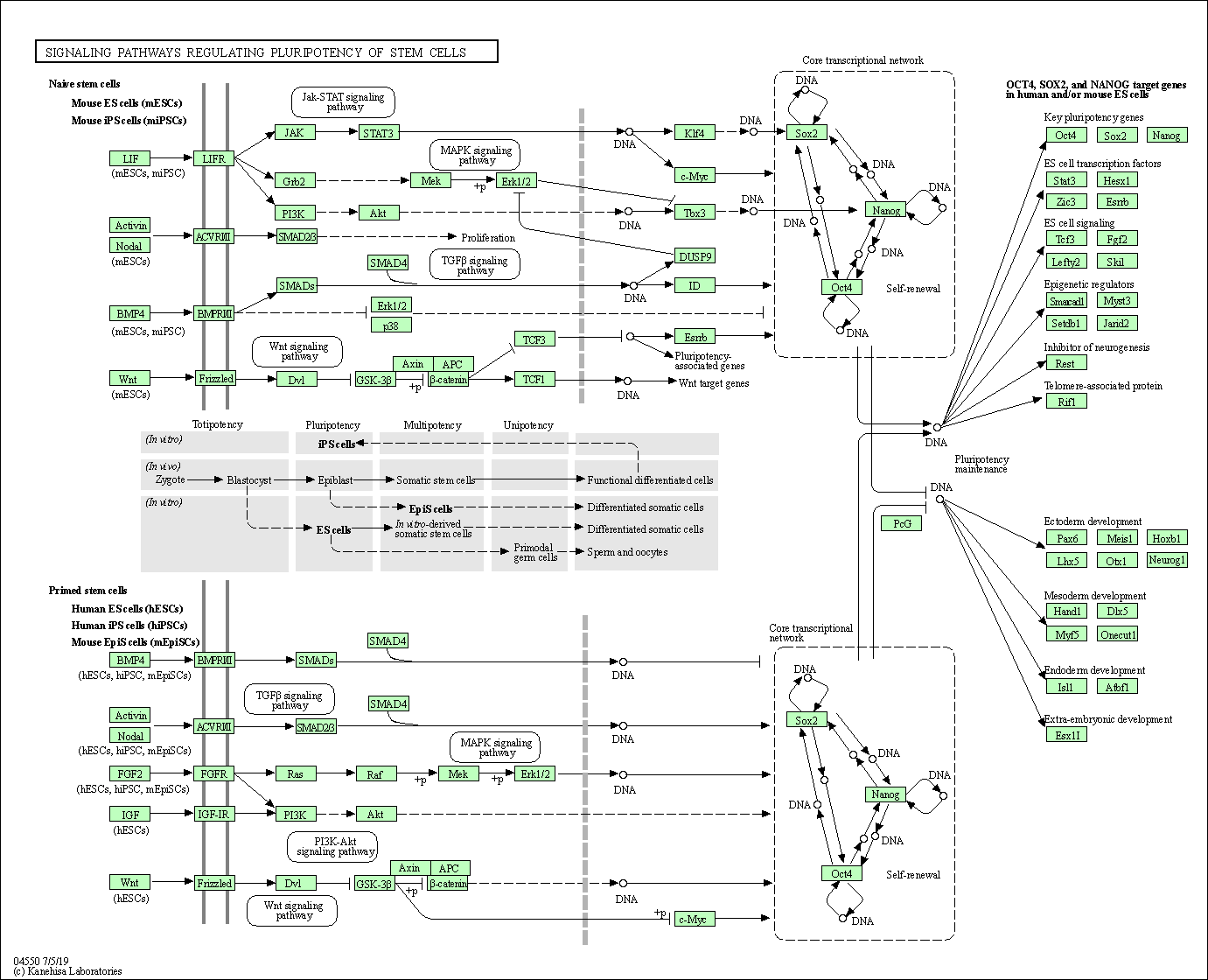
|
|||||||
| Vascular smooth muscle contraction | hsa04270 |
Pathway Map 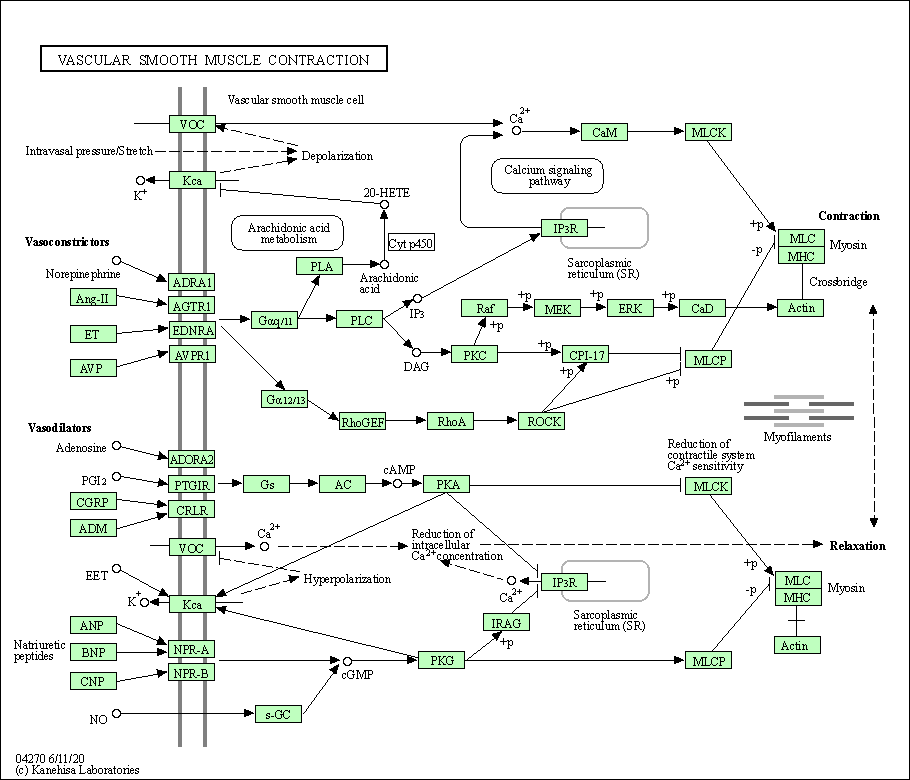
|
|||||||
| Cushing syndrome | hsa04934 |
Pathway Map 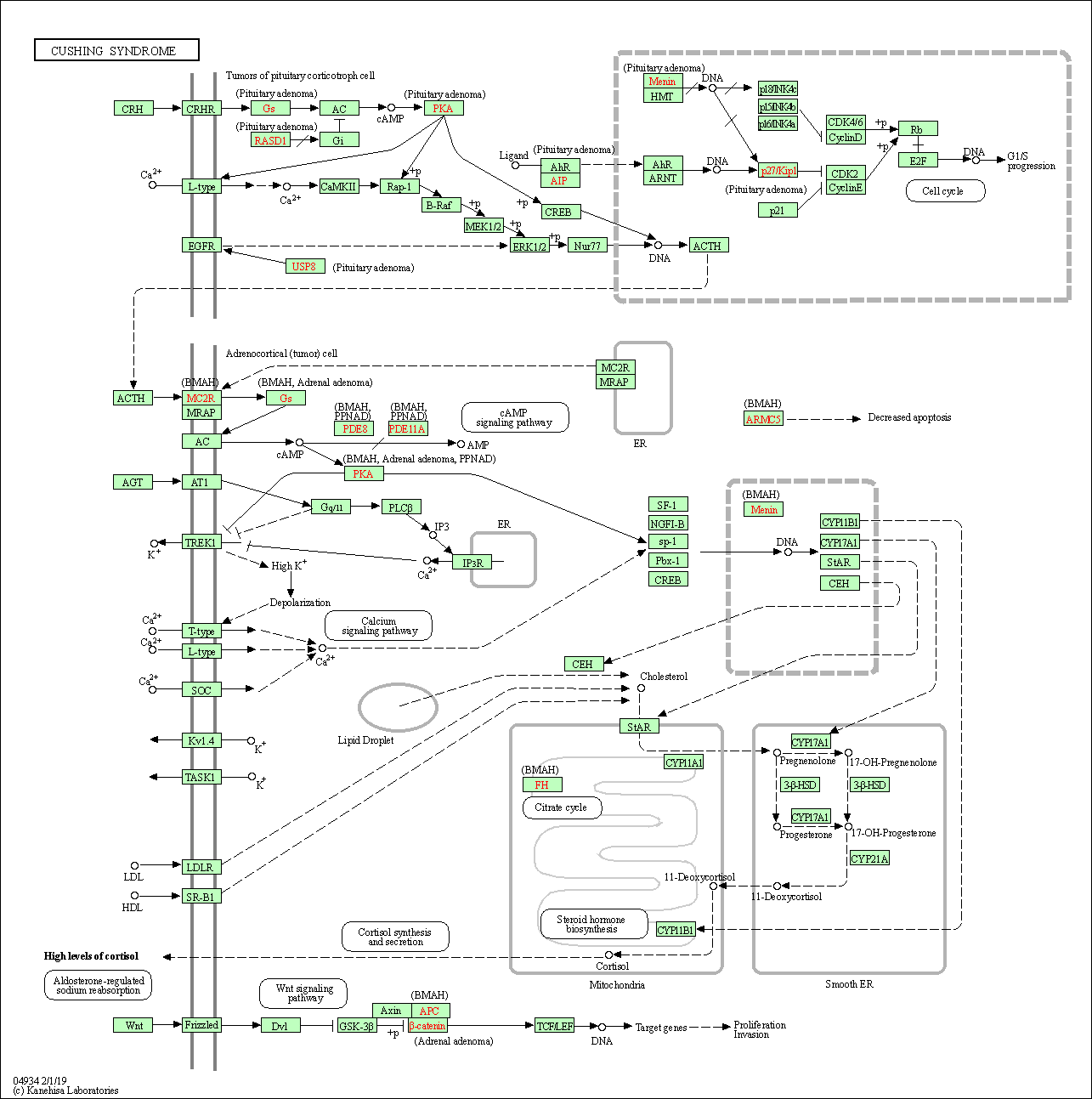
|
|||||||
| Insulin signaling pathway | hsa04910 |
Pathway Map 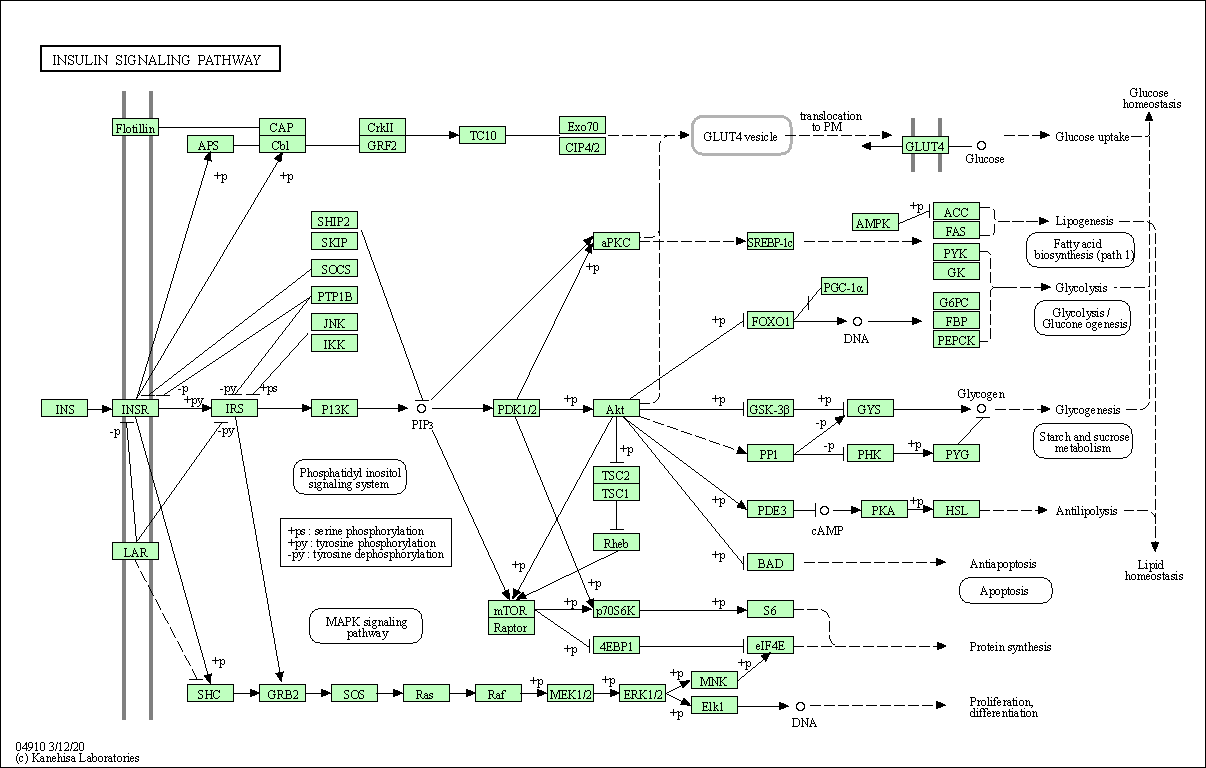
|
|||||||
| GnRH signaling pathway | hsa04912 |
Pathway Map 
|
|||||||
| Estrogen signaling pathway | hsa04915 |
Pathway Map 
|
|||||||
| Melanogenesis | hsa04916 |
Pathway Map 
|
|||||||
| Prolactin signaling pathway | hsa04917 |
Pathway Map 
|
|||||||
| Thyroid hormone signaling pathway | hsa04919 |
Pathway Map 
|
|||||||
| Oxytocin signaling pathway | hsa04921 |
Pathway Map 
|
|||||||
| Relaxin signaling pathway | hsa04926 |
Pathway Map 
|
|||||||
| GnRH secretion | hsa04929 |
Pathway Map 
|
|||||||
| Growth hormone synthesis, secretion and action | hsa04935 |
Pathway Map 
|
|||||||
| Long-term potentiation | hsa04720 |
Pathway Map 
|
|||||||
| Neurotrophin signaling pathway | hsa04722 |
Pathway Map 
|
|||||||
| Long-term depression | hsa04730 |
Pathway Map 
|
|||||||
| Alzheimer disease | hsa05010 |
Pathway Map 
|
|||||||
| Pathways of neurodegeneration - multiple diseases | hsa05022 |
Pathway Map 
|
|||||||
| MAPK signaling pathway | hsa04010 |
Pathway Map 
|
|||||||
| ErbB signaling pathway | hsa04012 |
Pathway Map 
|
|||||||
| Ras signaling pathway | hsa04014 |
Pathway Map 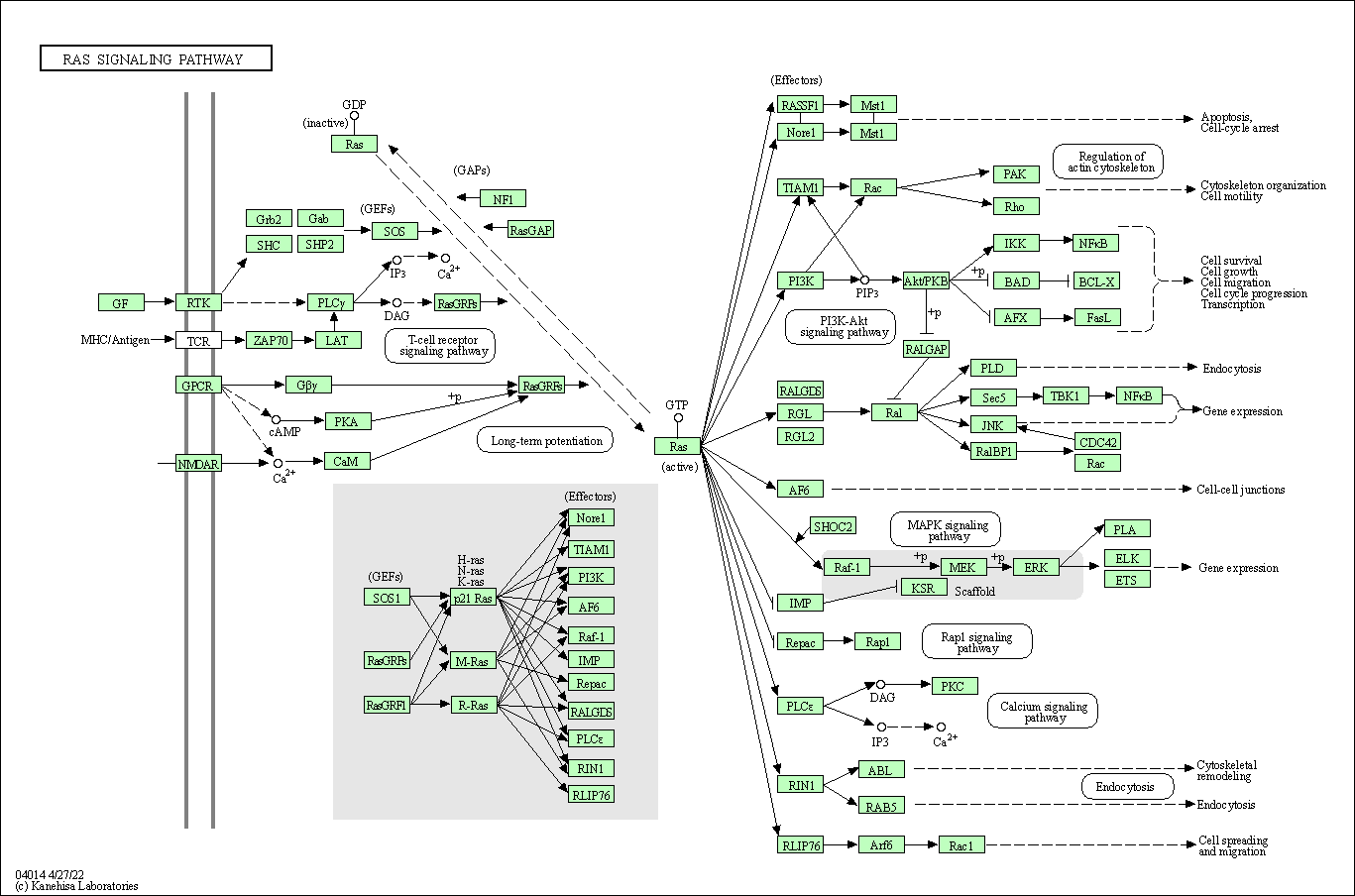
|
|||||||
| Rap1 signaling pathway | hsa04015 |
Pathway Map 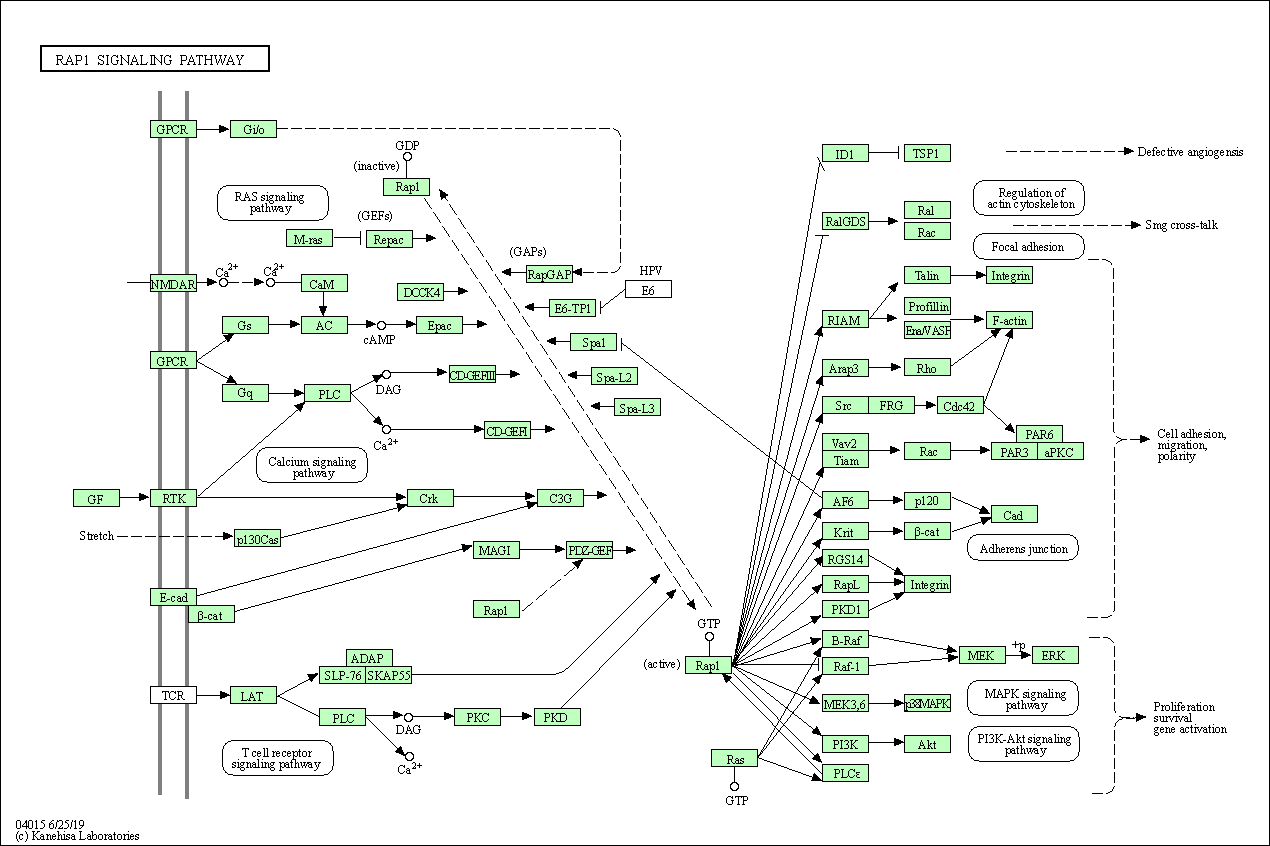
|
|||||||
| cGMP-PKG signaling pathway | hsa04022 |
Pathway Map 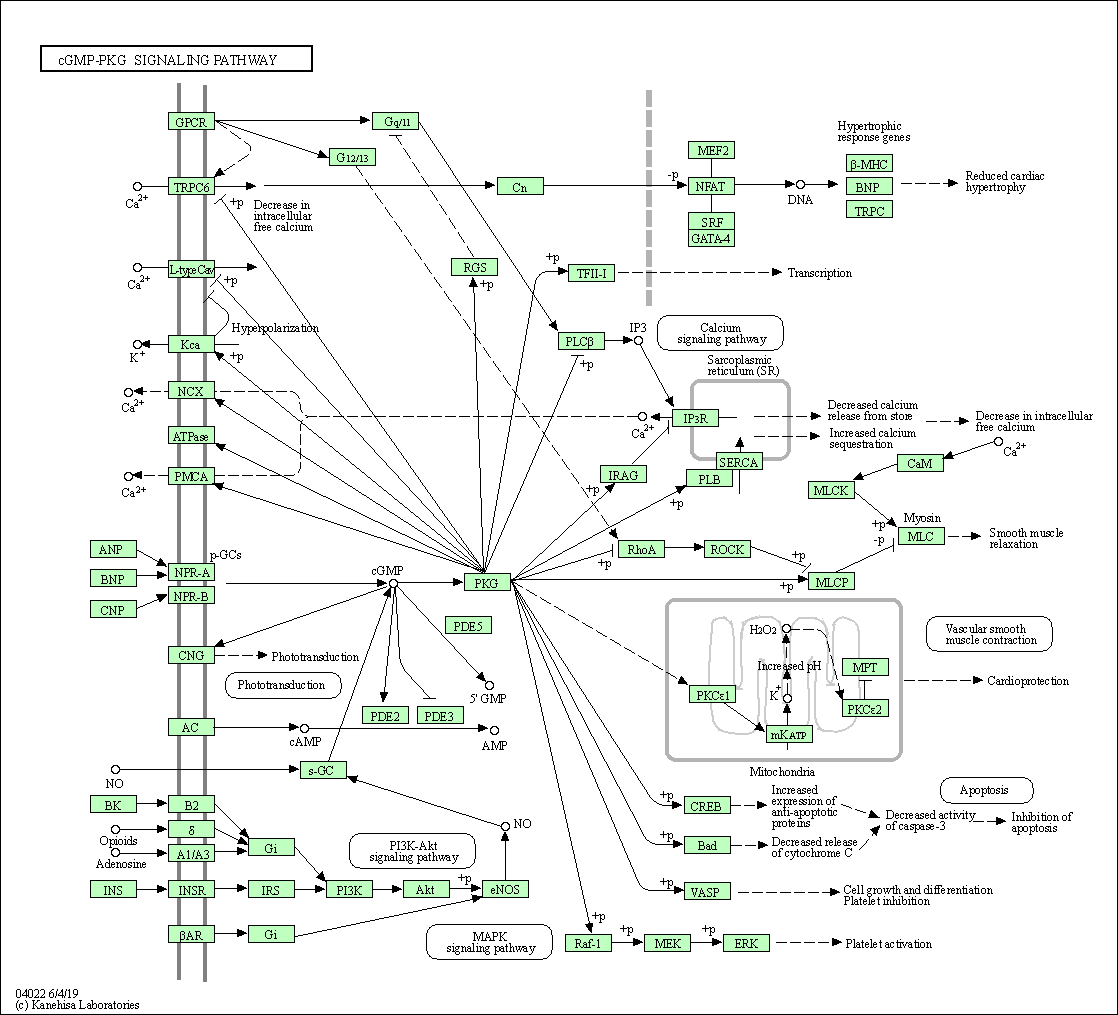
|
|||||||
| cAMP signaling pathway | hsa04024 |
Pathway Map 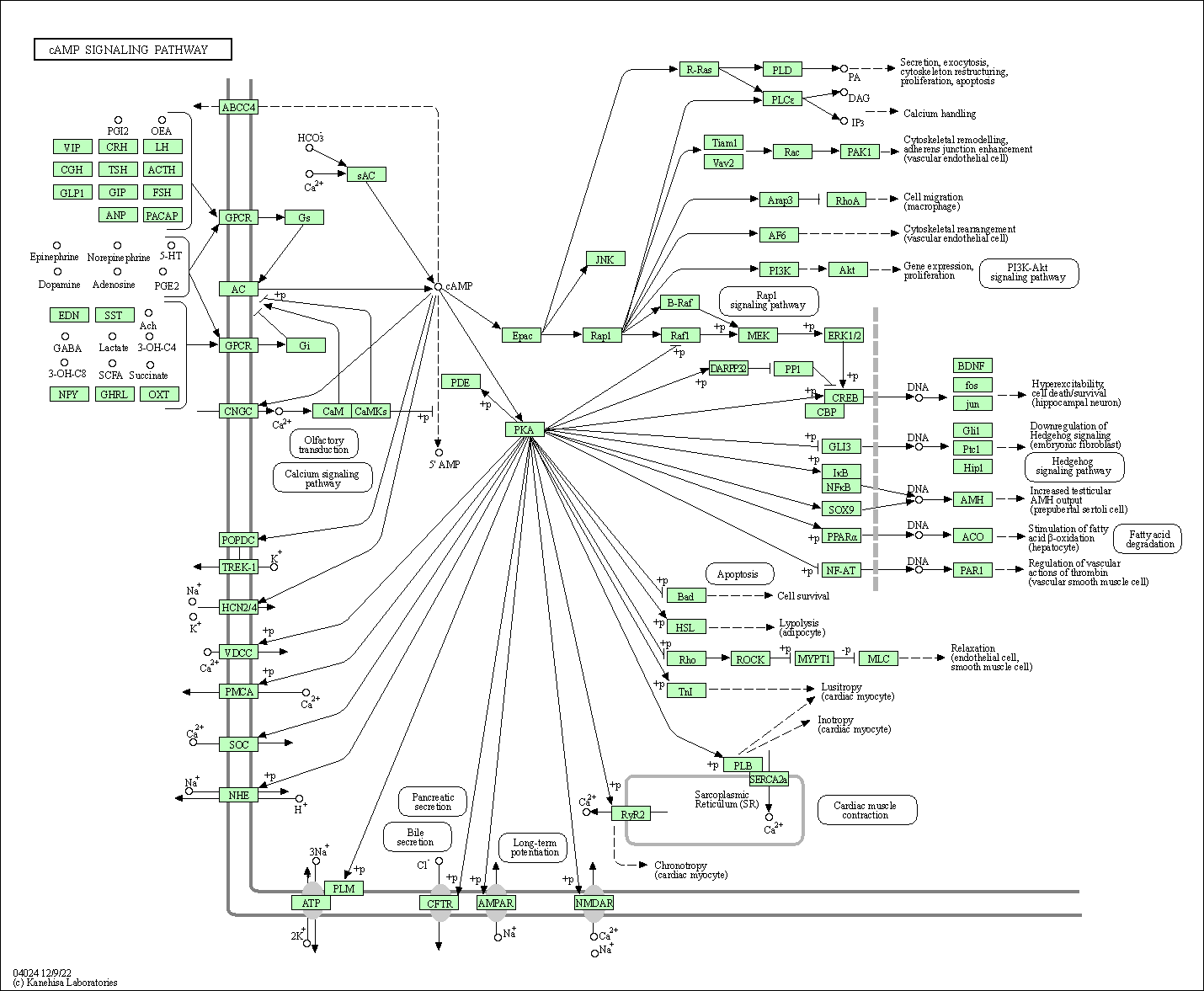
|
|||||||
| HIF-1 signaling pathway | hsa04066 |
Pathway Map 
|
|||||||
| FoxO signaling pathway | hsa04068 |
Pathway Map 
|
|||||||
| Sphingolipid signaling pathway | hsa04071 |
Pathway Map 
|
|||||||
| Phospholipase D signaling pathway | hsa04072 |
Pathway Map 
|
|||||||
| mTOR signaling pathway | hsa04150 |
Pathway Map 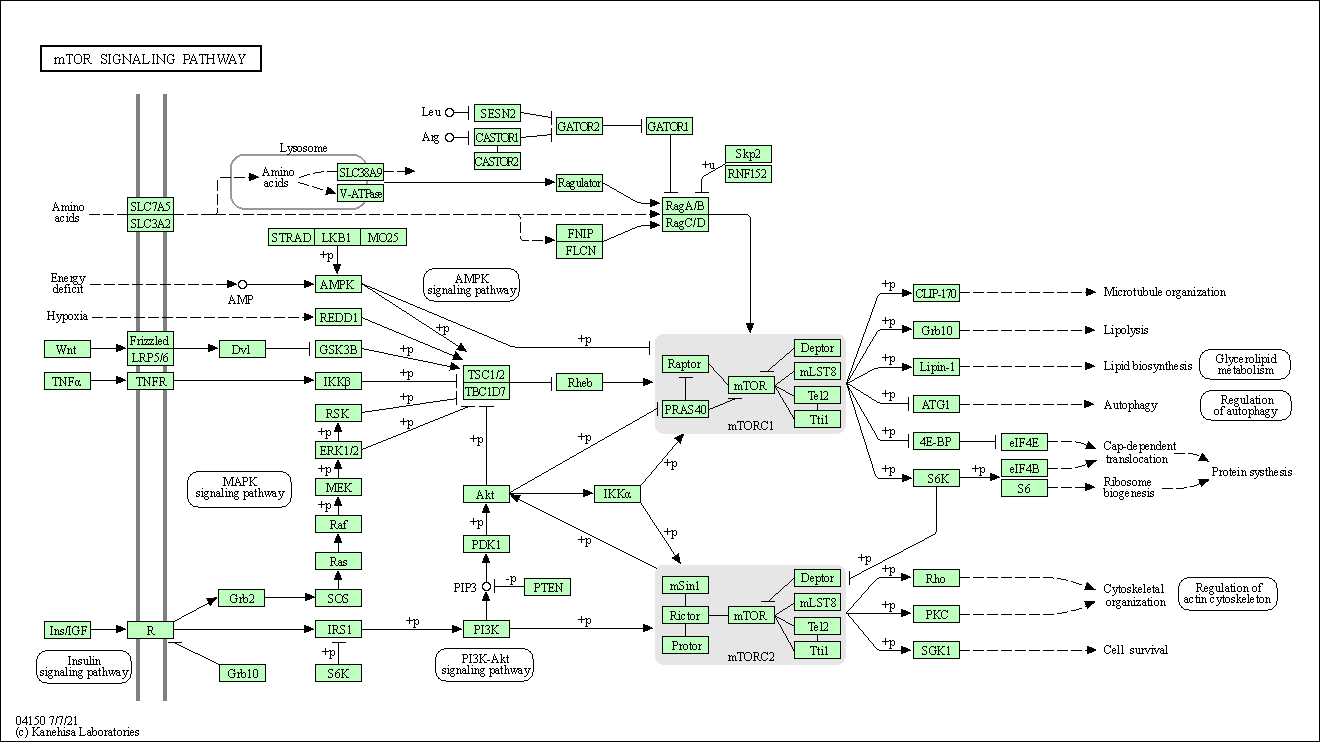
|
|||||||
| PI3K-Akt signaling pathway | hsa04151 |
Pathway Map 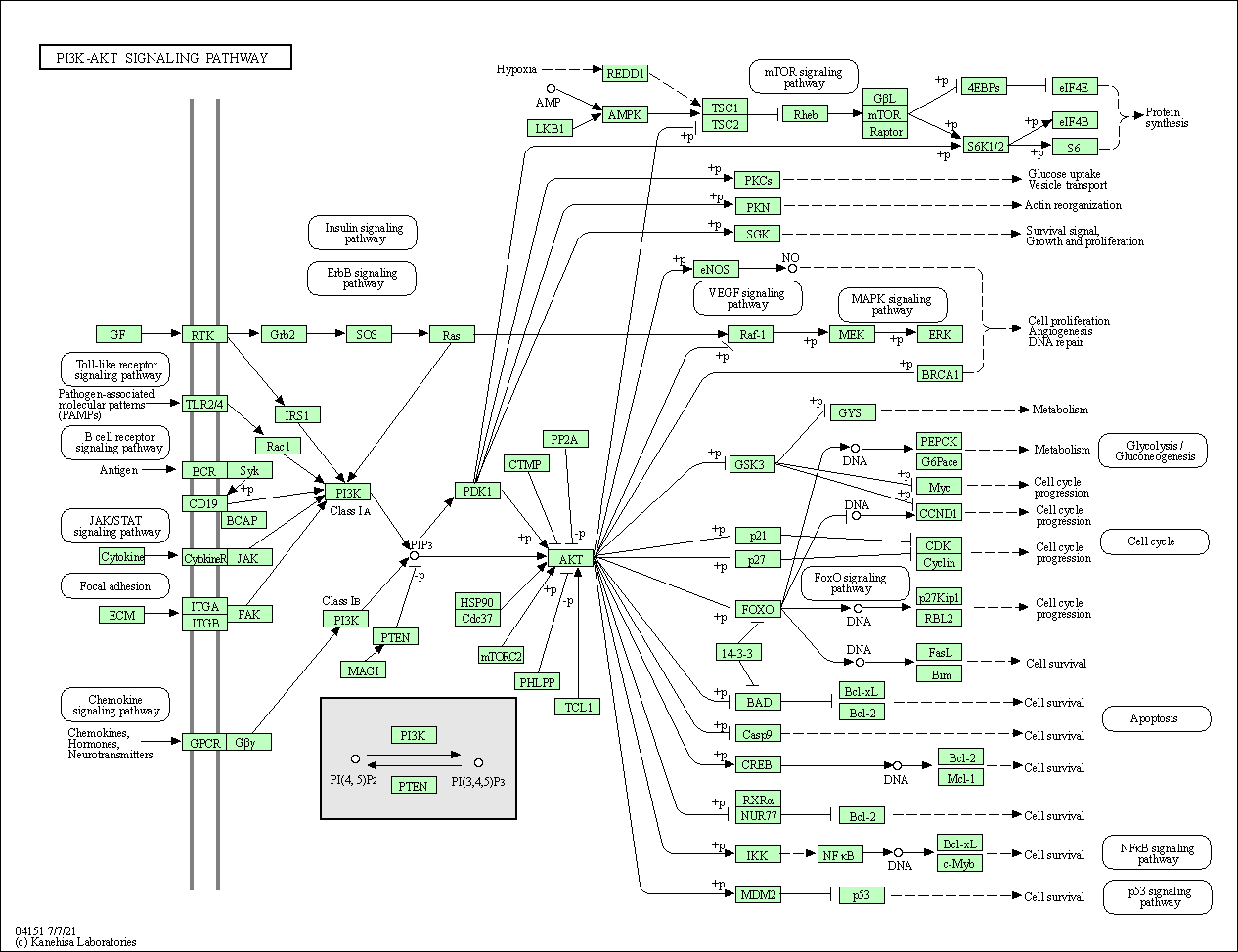
|
|||||||
| VEGF signaling pathway | hsa04370 |
Pathway Map 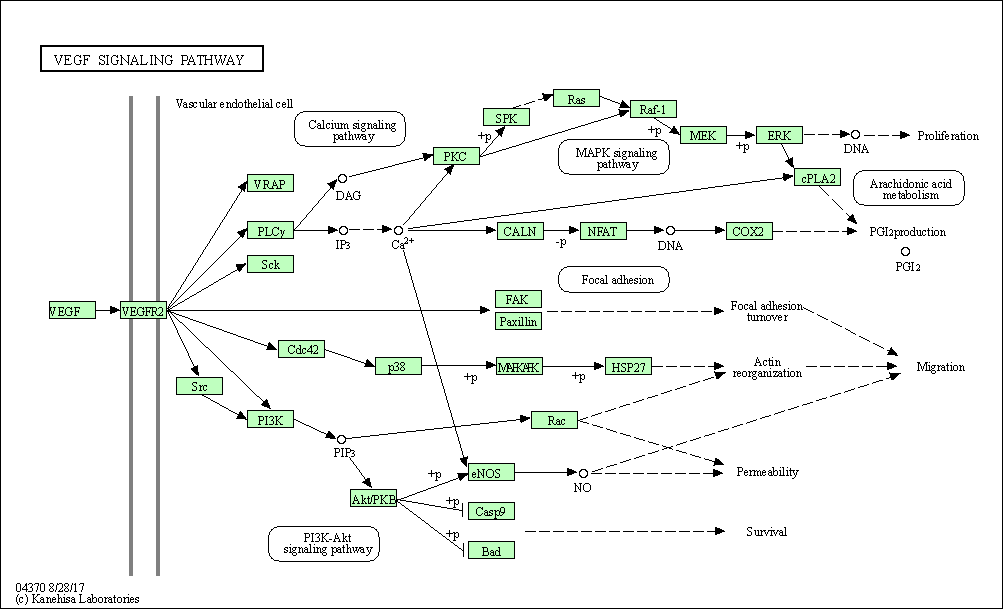
|
|||||||
| Apelin signaling pathway | hsa04371 |
Pathway Map 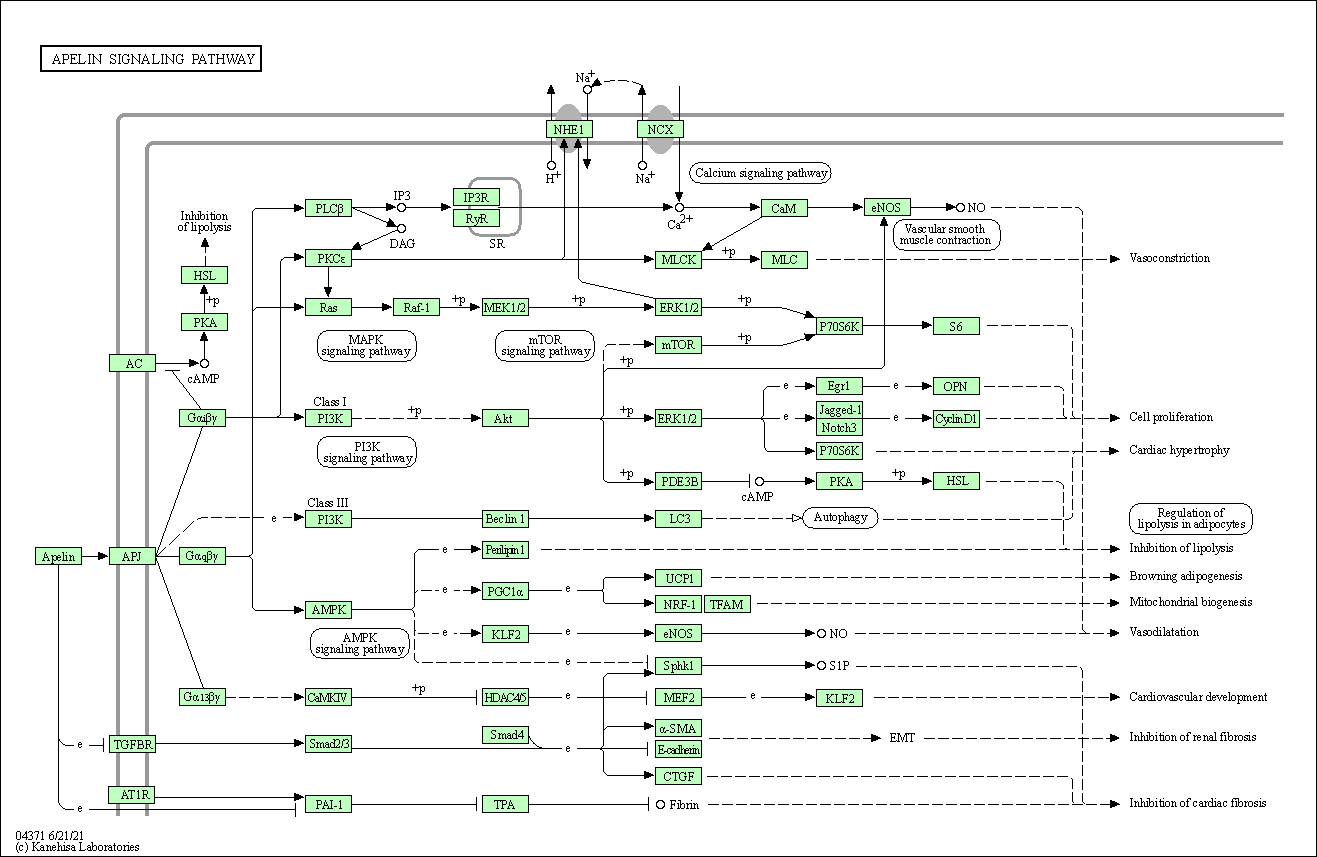
|
|||||||
| Autophagy - animal | hsa04140 |
Pathway Map 
|
|||||||
| 3D Structure |
|
||||||||
Full List of Virus RNA Interacting with This Protien
| Virus Name | Binding Region | RNA Info | Detection Method | Infection Cell | Cell ID | Cell Originated Tissue | Infection Time | Interaction Score 1 | Interaction Score 2 |
|---|---|---|---|---|---|---|---|---|---|
| Encephalomyocarditis virus | 5'UTR - 3'UTR | RNA Info | Comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS) | HuH-7.5 Cells (Human hepatocellular carcinoma cell) | . | Liver | . | Z-score = -2.122183279 | FZR = 1 |
| Influenza A virus | 5'UTR - 3'UTR | RNA Info | Comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS) | HuH-7.5 Cells (Human hepatocellular carcinoma cell) | . | Liver | . | Z-score = -3.920753393 | FZR = 0.076267493 |
Potential Drug(s) that Targets This Protein
Protein Sequence Information
|
MLARRKPVLPALTINPTIAEGPSPTSEGASEANLVDLQKKLEELELDEQQKKRLEAFLTQKAKVGELKDDDFERISELGAGNGGVVTKVQHRPSGLIMARKLIHLEIKPAIRNQIIRELQVLHECNSPYIVGFYGAFYSDGEISICMEHMDGGSLDQVLKEAKRIPEEILGKVSIAVLRGLAYLREKHQIMHRDVKPSNILVNSRGEIKLCDFGVSGQLIDSMANSFVGTRSYMAPERLQGTHYSVQSDIWSMGLSLVELAVGRYPIPPPDAKELEAIFGRPVVDGEEGEPHSISPRPRPPGRPVSGHGMDSRPAMAIFELLDYIVNEPPPKLPNGVFTPDFQEFVNKCLIKNPAERADLKMLTNHTFIKRSEVEEVDFAGWLCKTLRLNQPGTPTRTAV
Click to Show/Hide
|
