Host Protein General Information
| Protein Name |
PP-1B
|
Gene Name |
PPP1CB
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Host Species |
Homo sapiens
|
Uniprot Entry Name |
PP1B_HUMAN
|
||||||
| Protein Families |
PPP phosphatase family
|
||||||||
| EC Number |
3.1.3.16; 3.1.3.53
|
||||||||
| Subcellular Location |
Cytoplasm
|
||||||||
| External Link | |||||||||
| NCBI Gene ID | |||||||||
| Uniprot ID | |||||||||
| Ensembl ID | |||||||||
| HGNC ID | |||||||||
| Related KEGG Pathway | |||||||||
| Platelet activation | hsa04611 |
Pathway Map 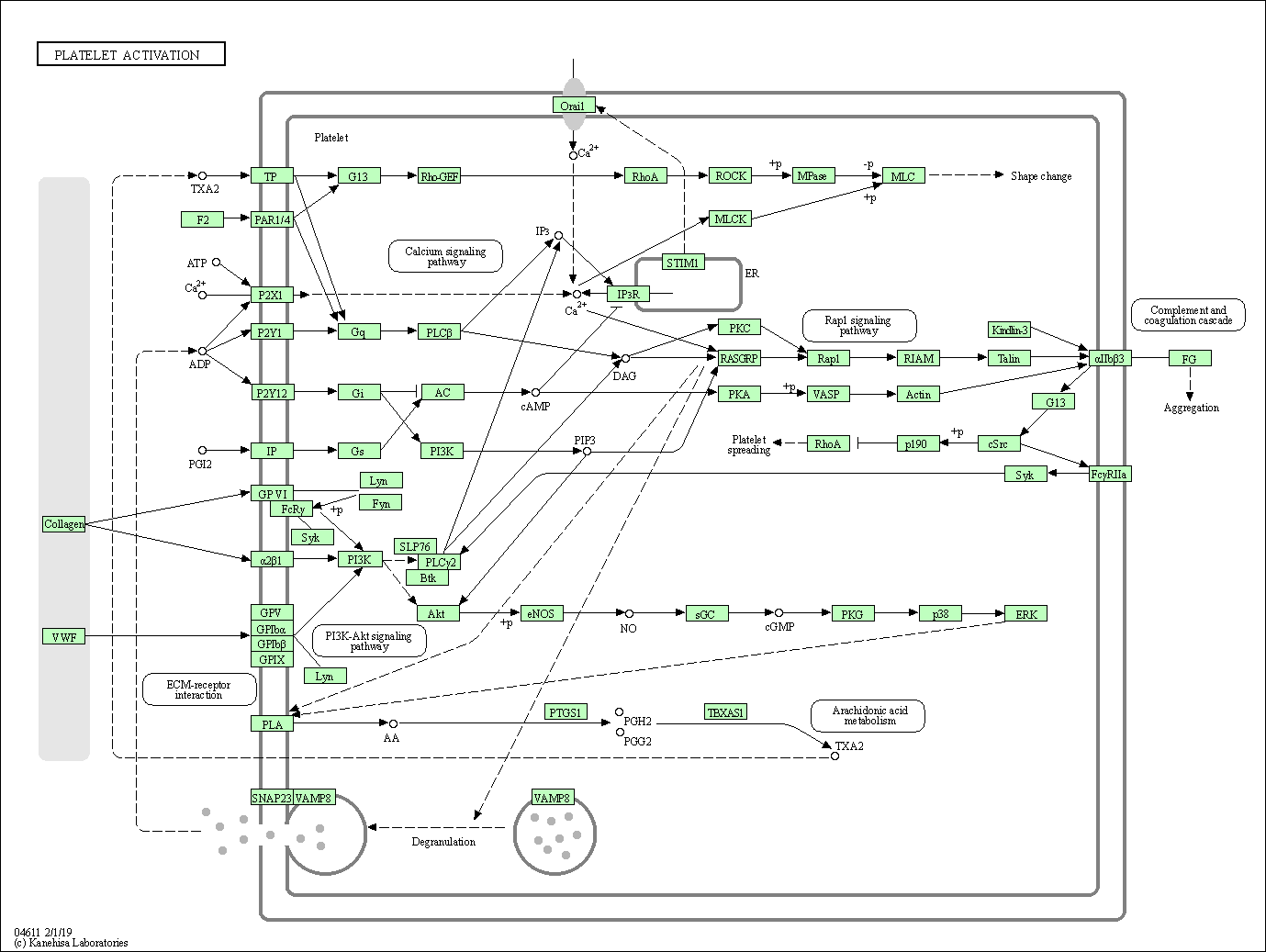
|
|||||||
| Herpes simplex virus 1 infection | hsa05168 |
Pathway Map 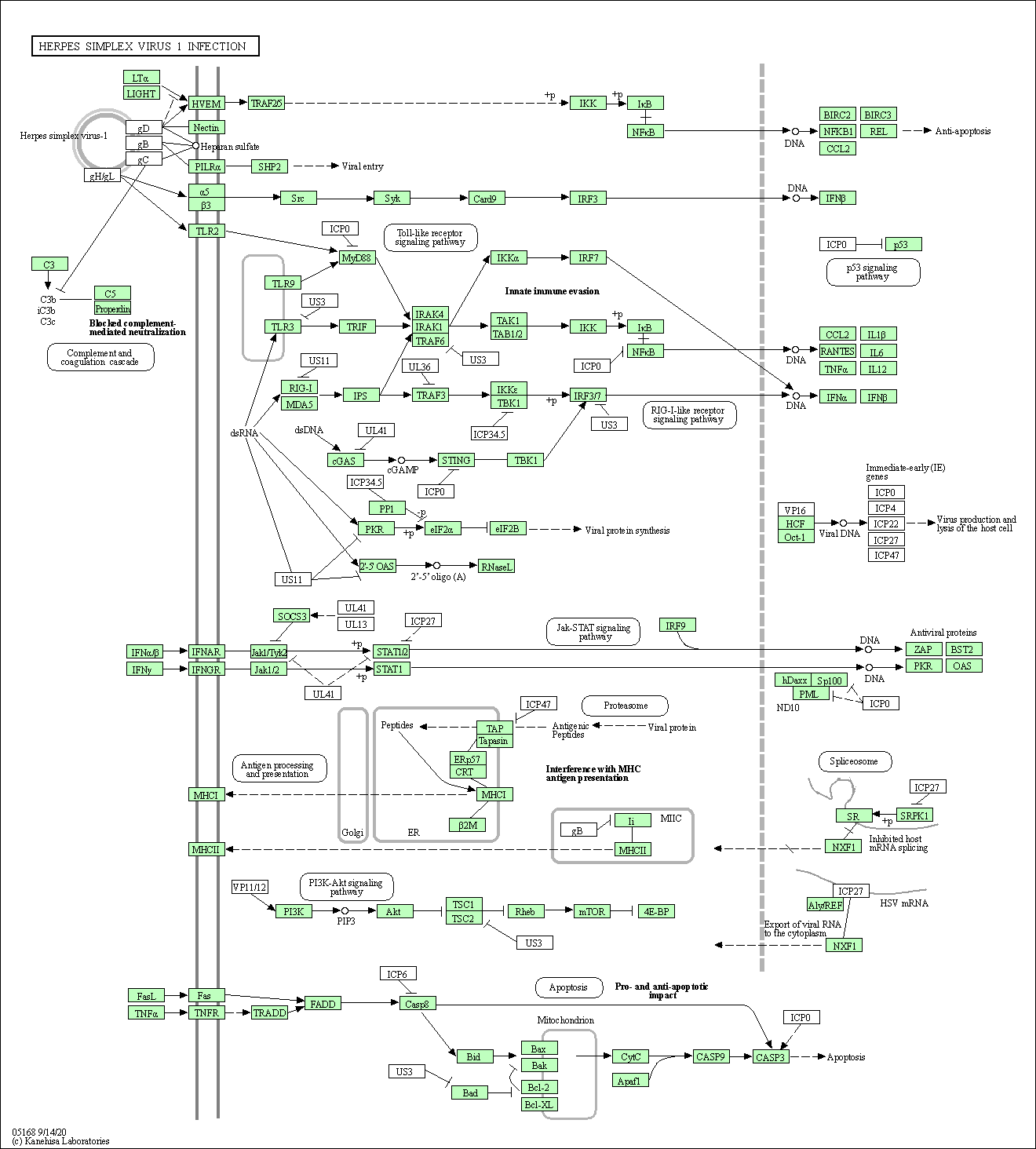
|
|||||||
| Proteoglycans in cancer | hsa05205 |
Pathway Map 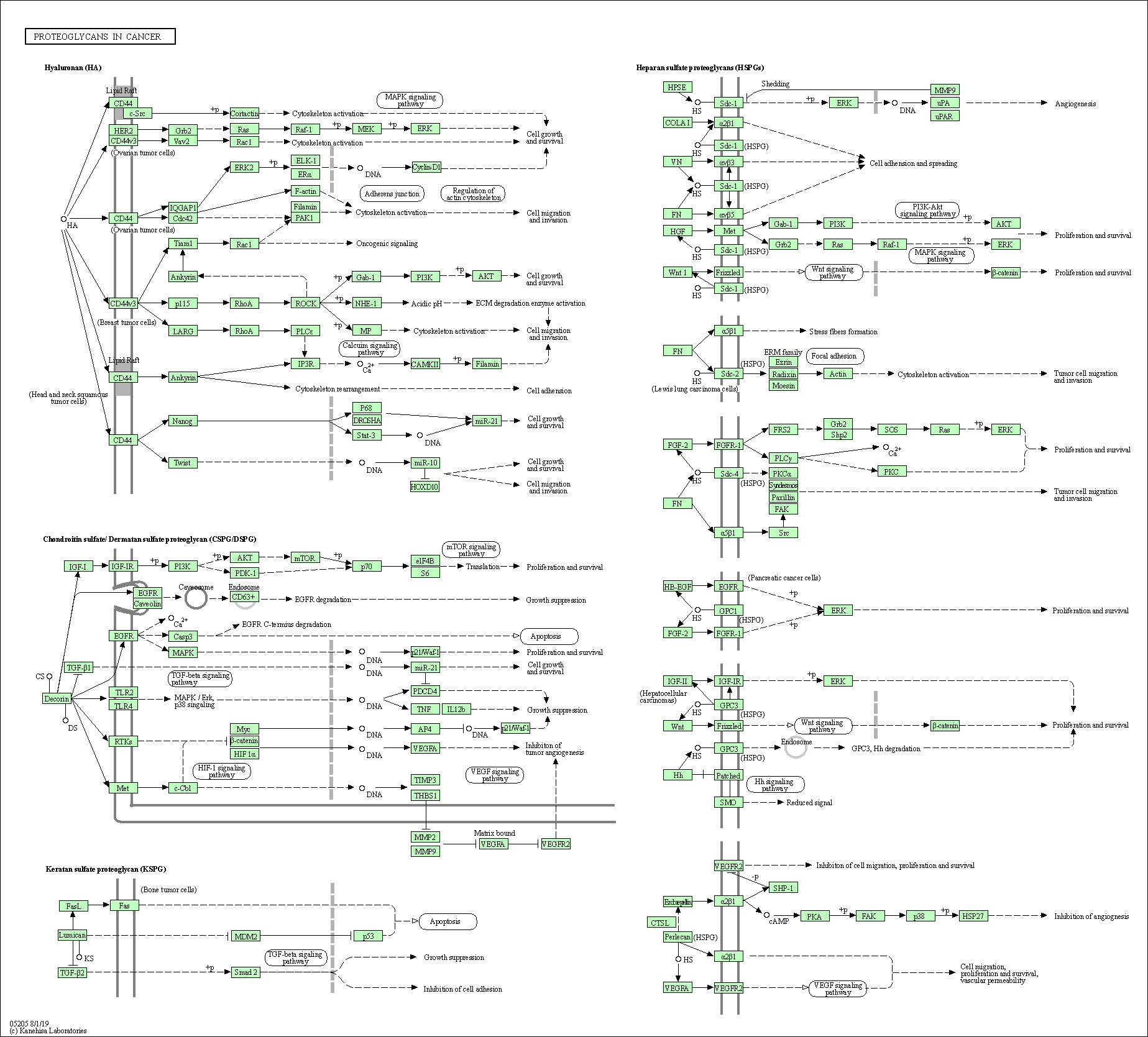
|
|||||||
| Diabetic cardiomyopathy | hsa05415 |
Pathway Map 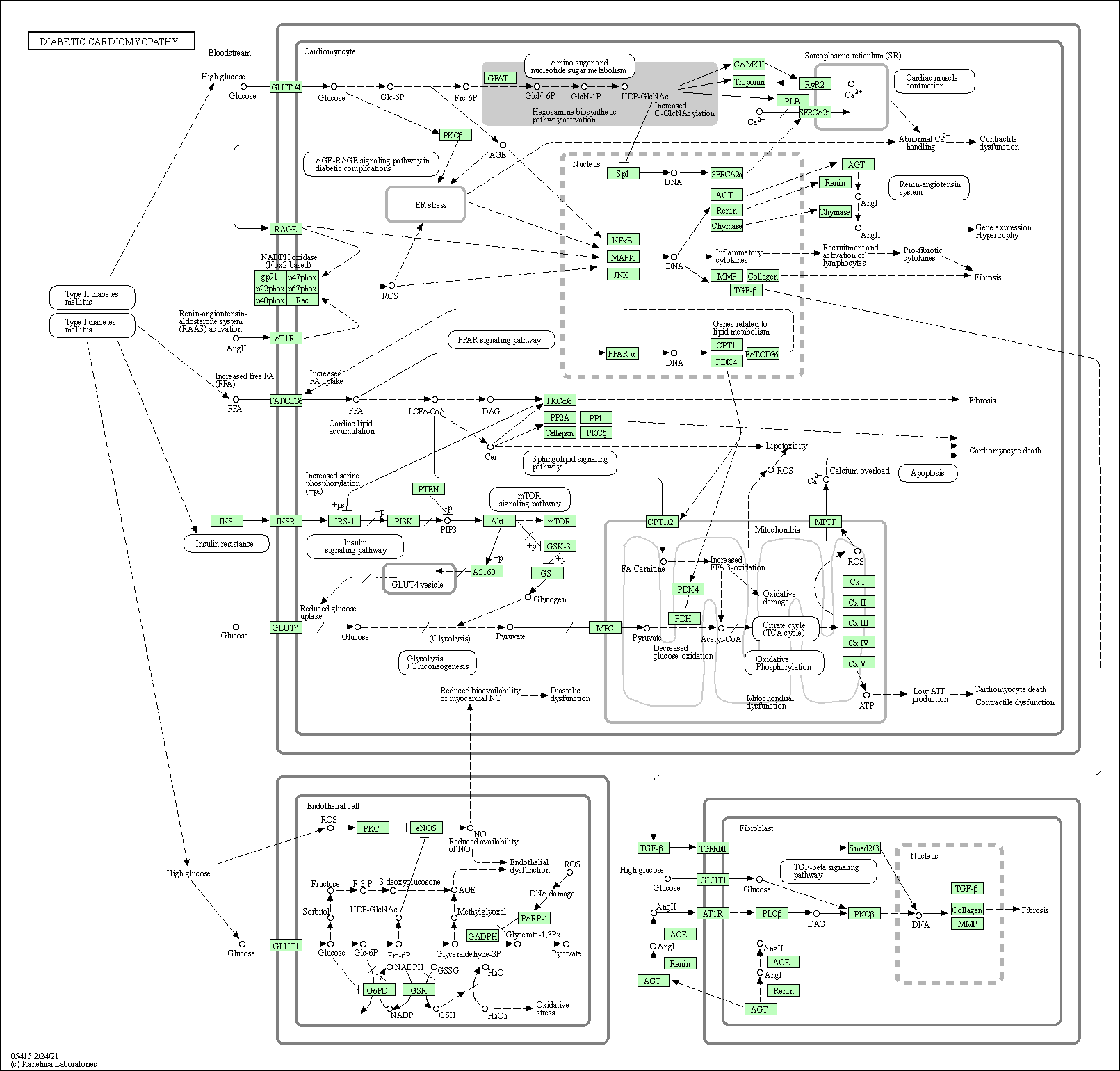
|
|||||||
| Oocyte meiosis | hsa04114 |
Pathway Map 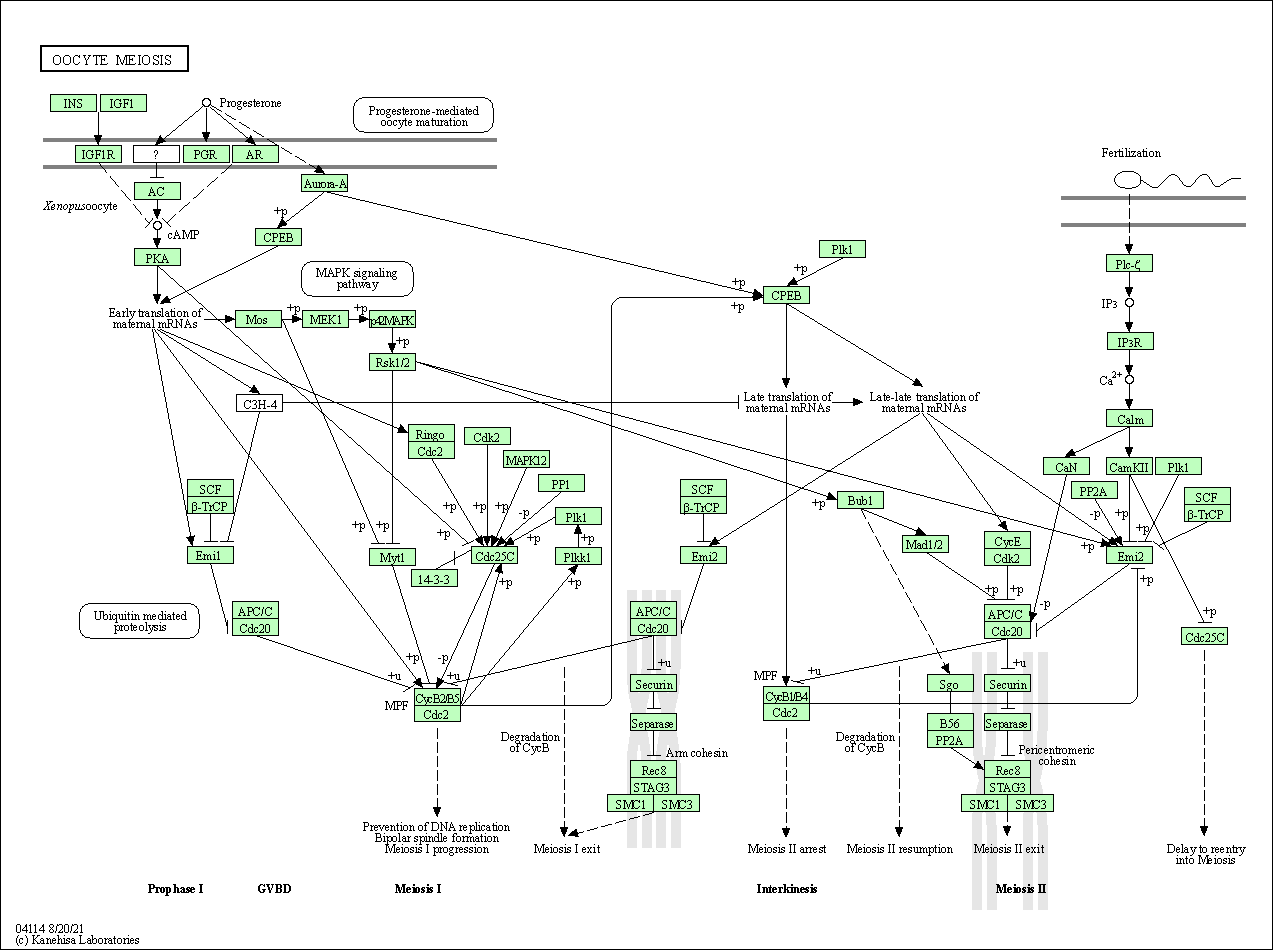
|
|||||||
| Cellular senescence | hsa04218 |
Pathway Map 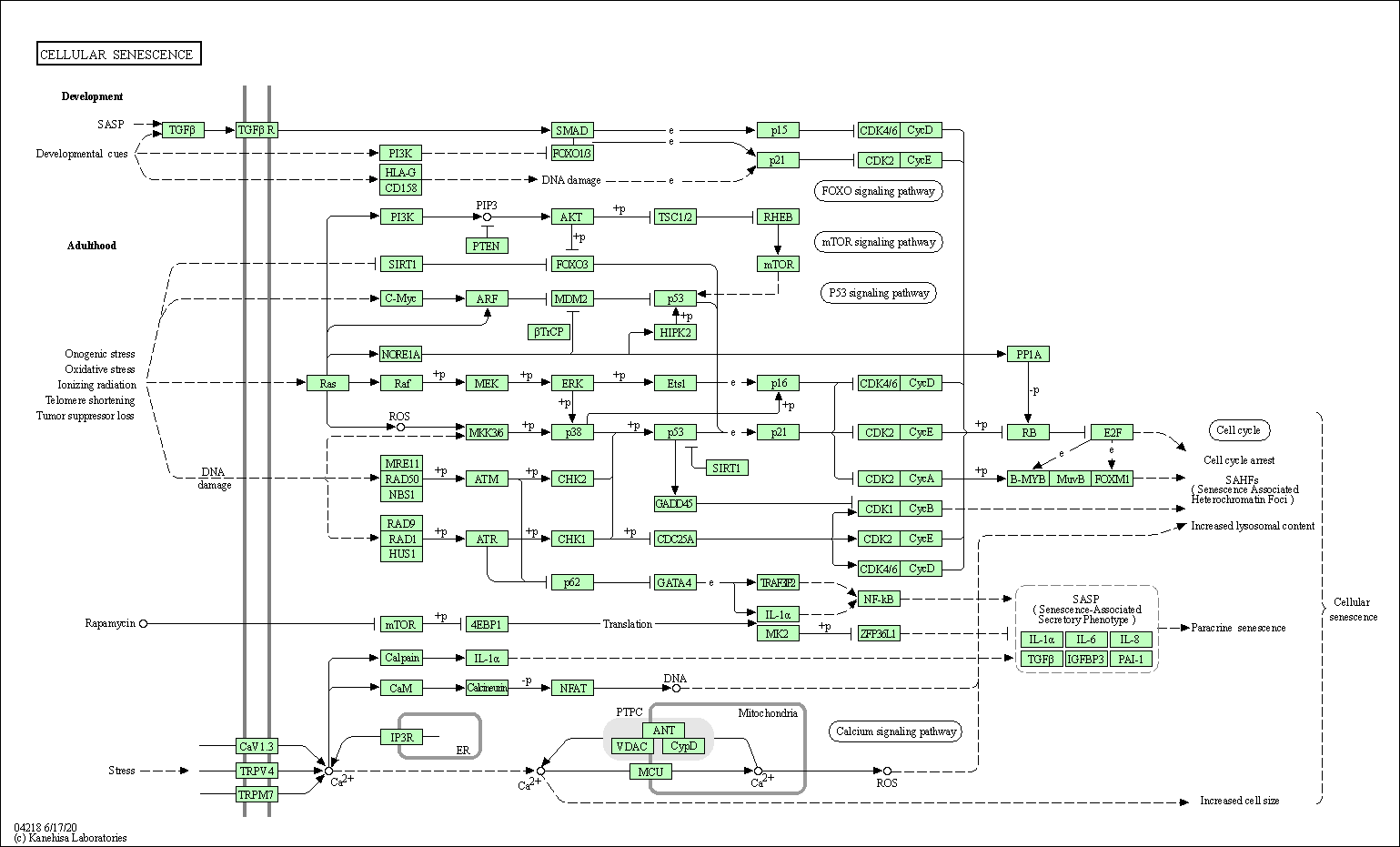
|
|||||||
| Regulation of actin cytoskeleton | hsa04810 |
Pathway Map 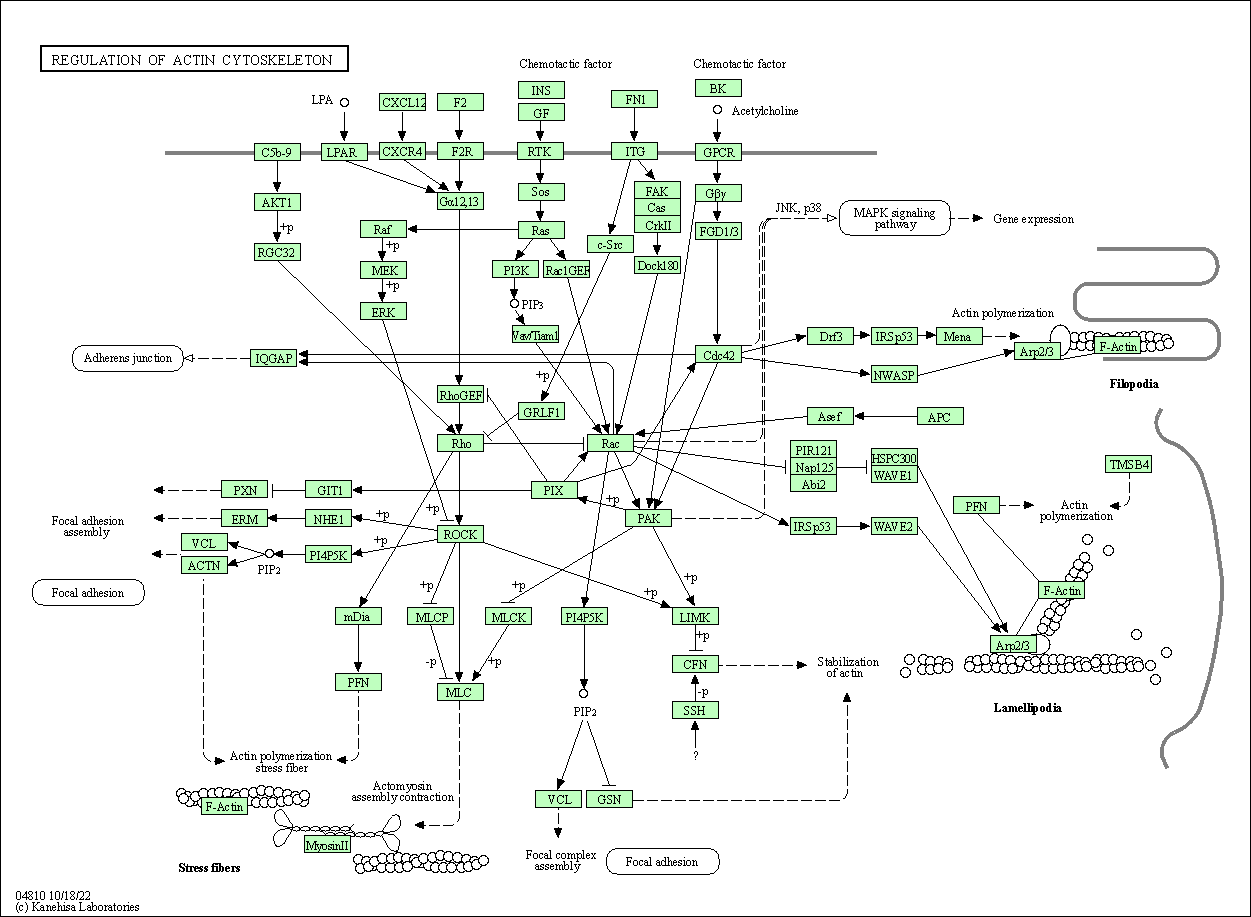
|
|||||||
| Focal adhesion | hsa04510 |
Pathway Map 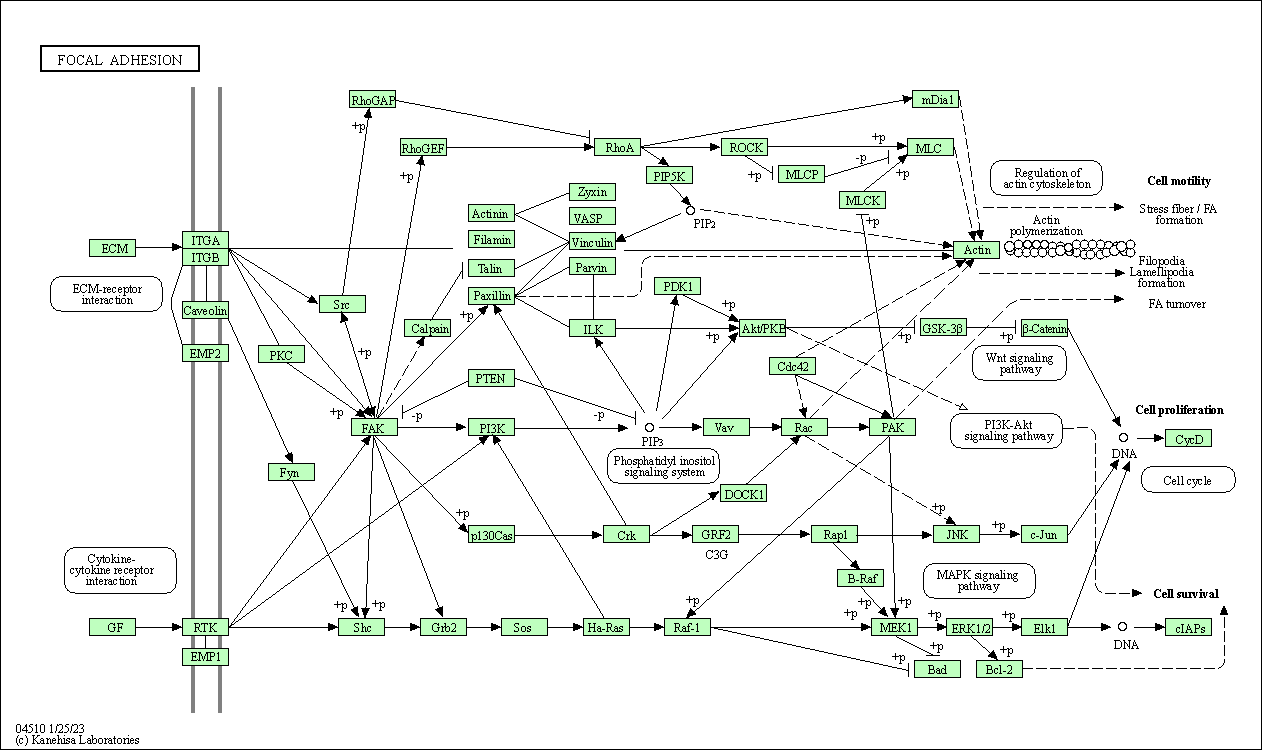
|
|||||||
| Adrenergic signaling in cardiomyocytes | hsa04261 |
Pathway Map 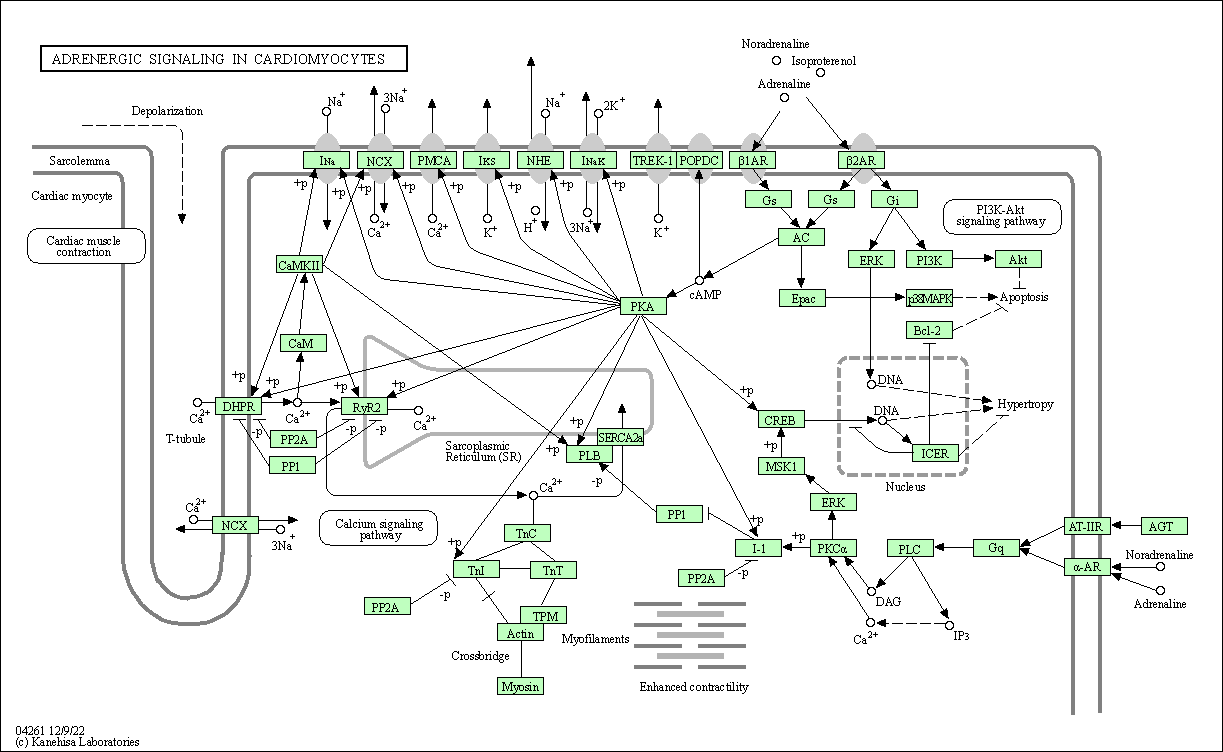
|
|||||||
| Vascular smooth muscle contraction | hsa04270 |
Pathway Map 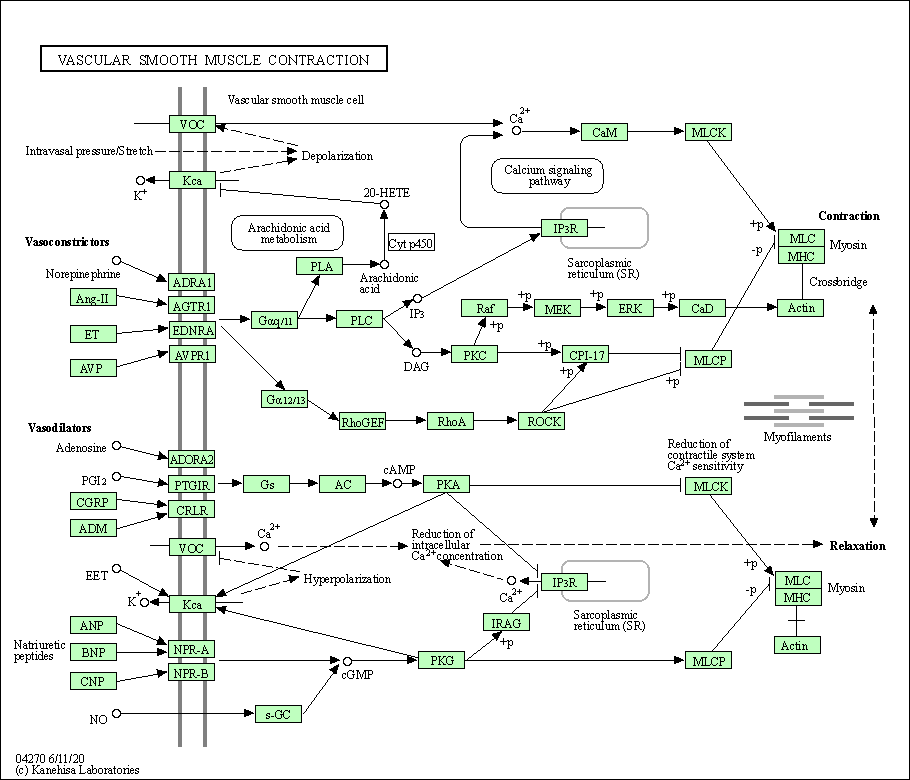
|
|||||||
| Insulin resistance | hsa04931 |
Pathway Map 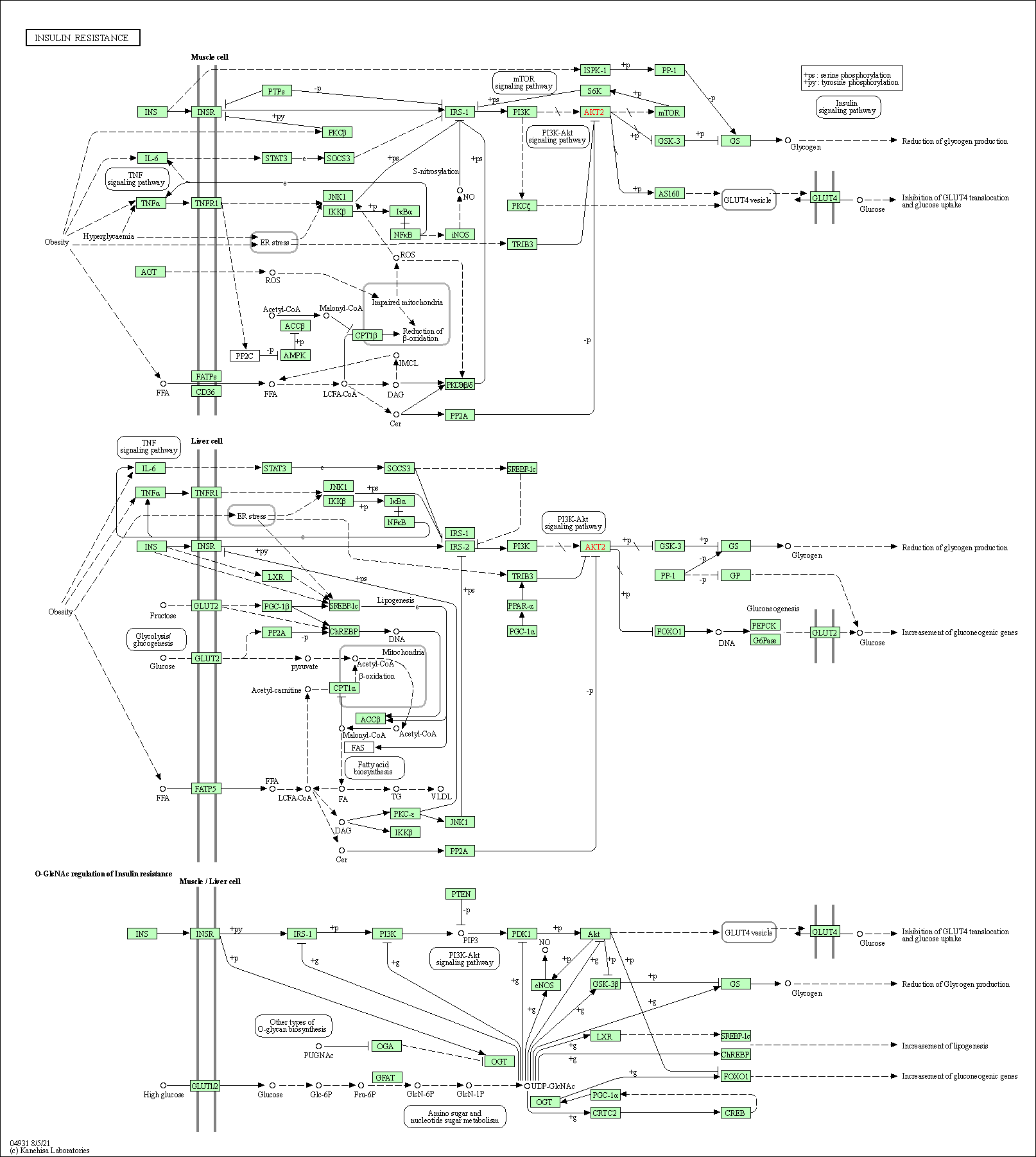
|
|||||||
| Insulin signaling pathway | hsa04910 |
Pathway Map 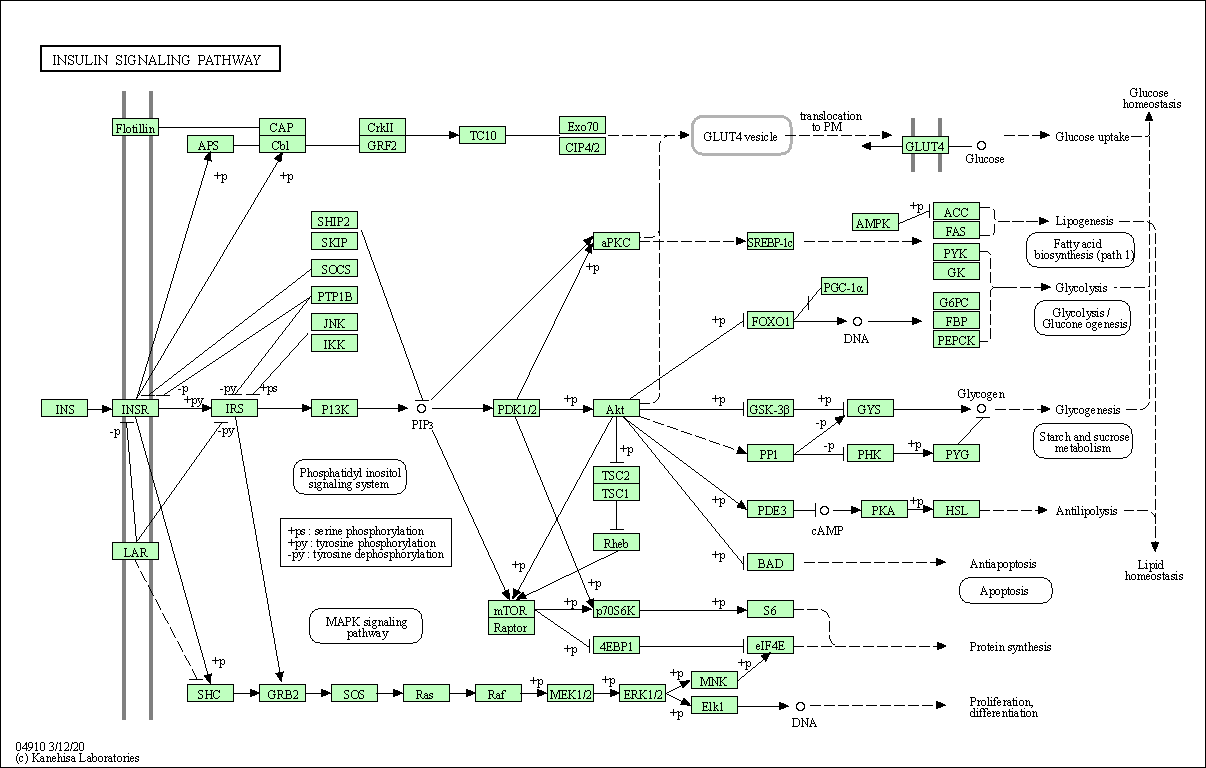
|
|||||||
| Oxytocin signaling pathway | hsa04921 |
Pathway Map 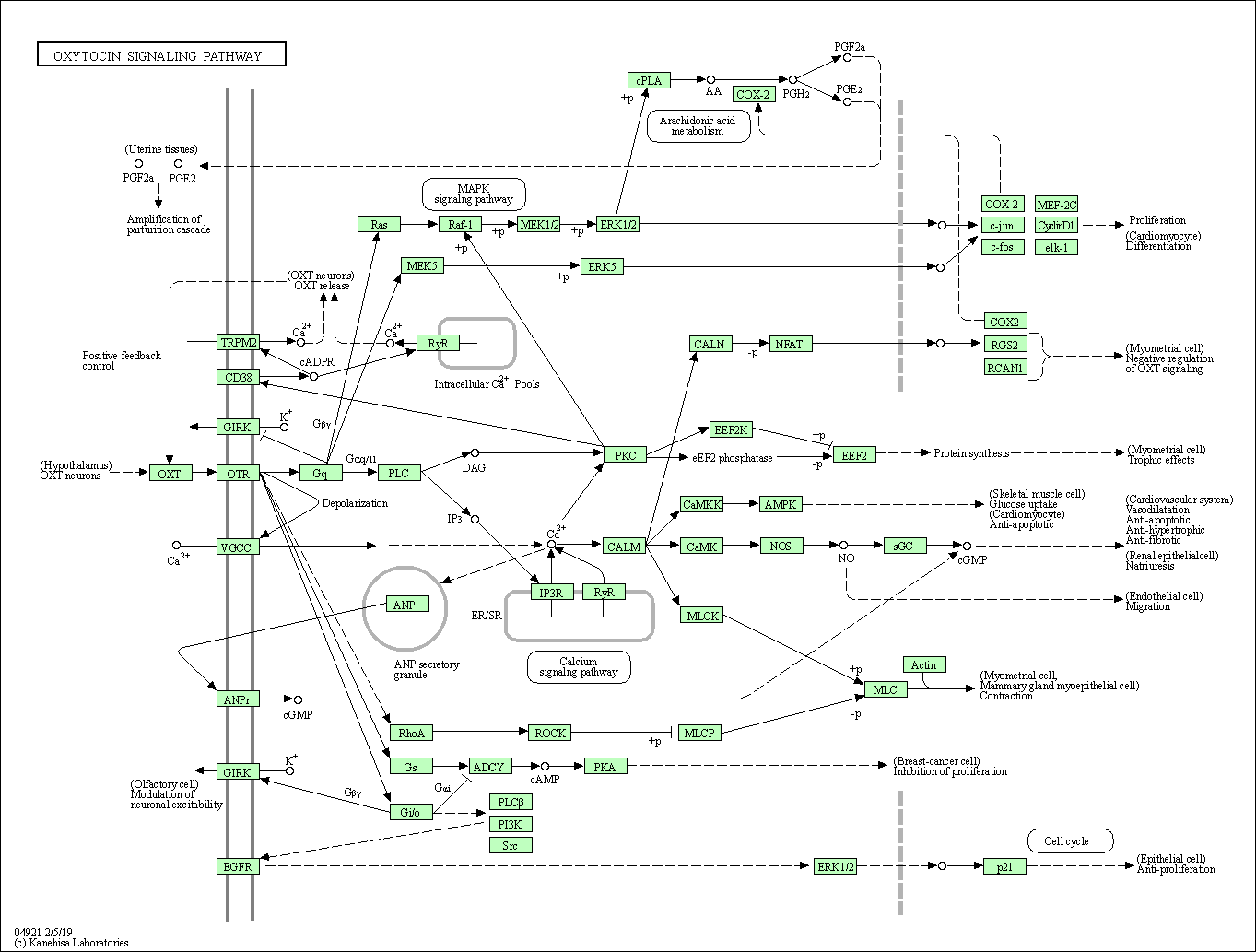
|
|||||||
| Long-term potentiation | hsa04720 |
Pathway Map 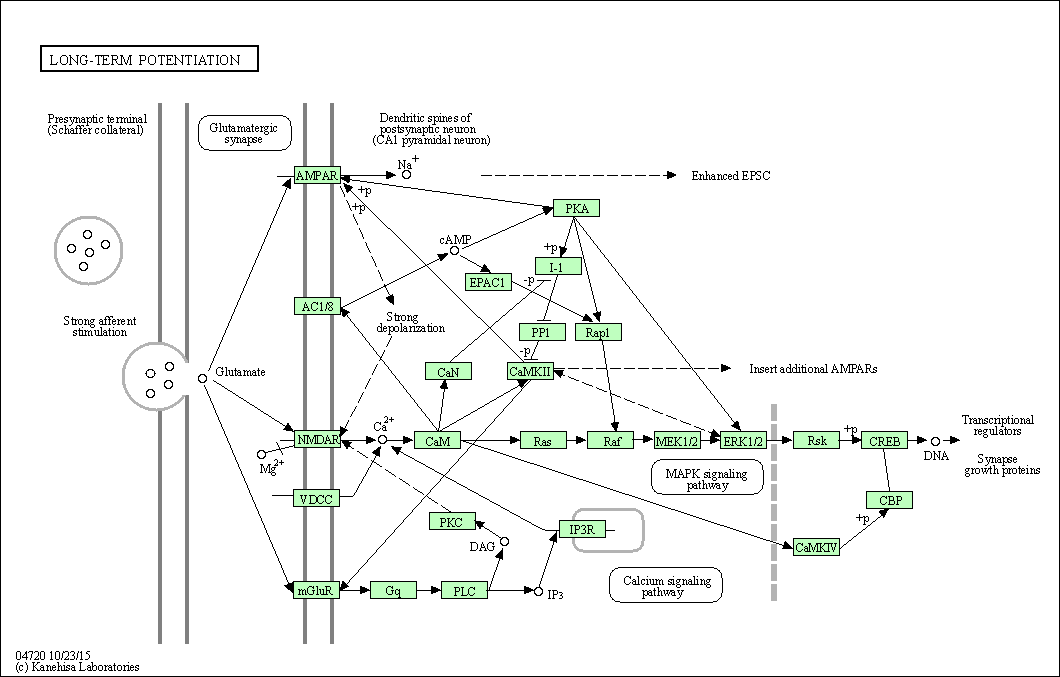
|
|||||||
| Dopaminergic synapse | hsa04728 |
Pathway Map 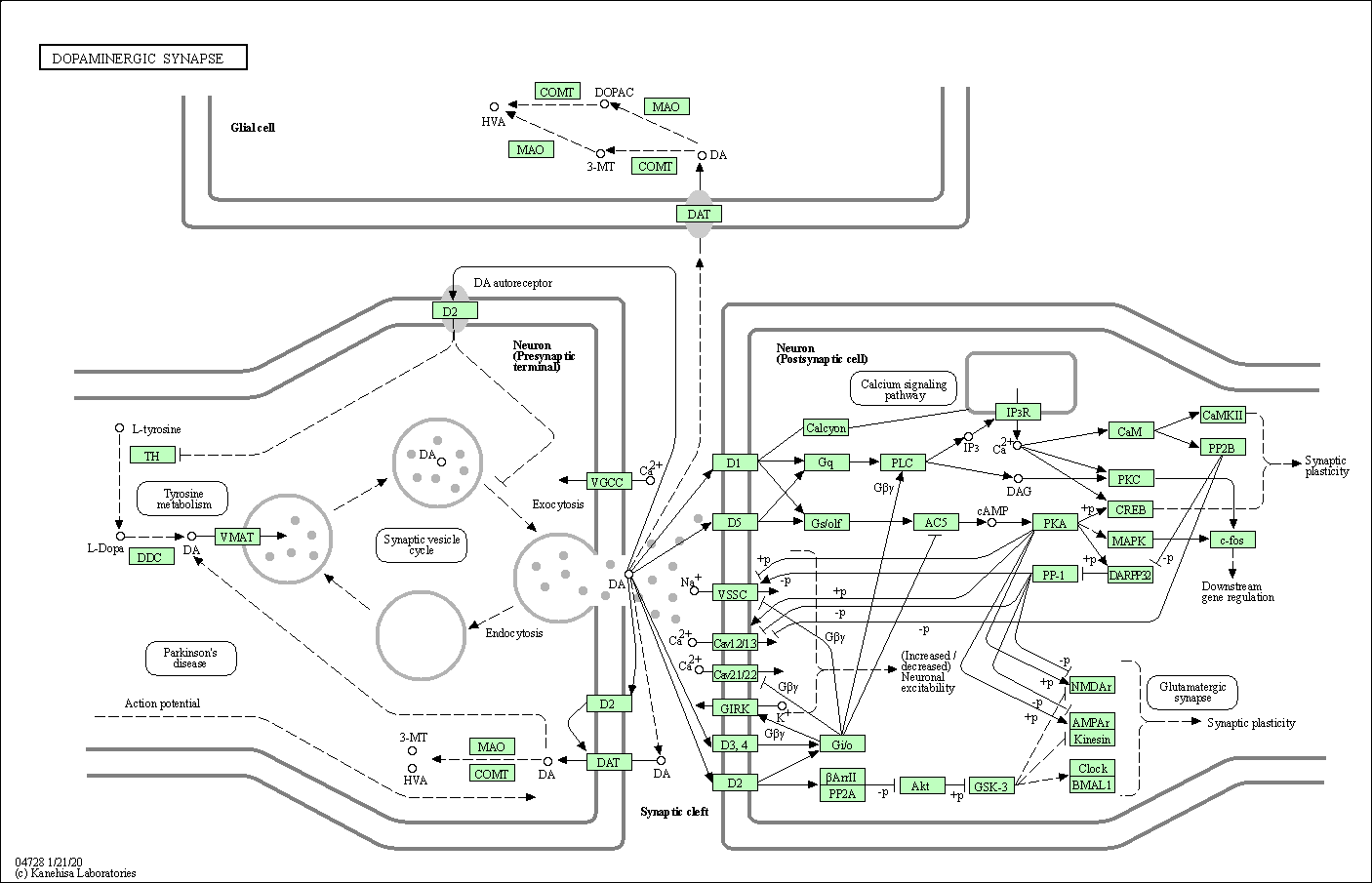
|
|||||||
| Inflammatory mediator regulation of TRP channels | hsa04750 |
Pathway Map 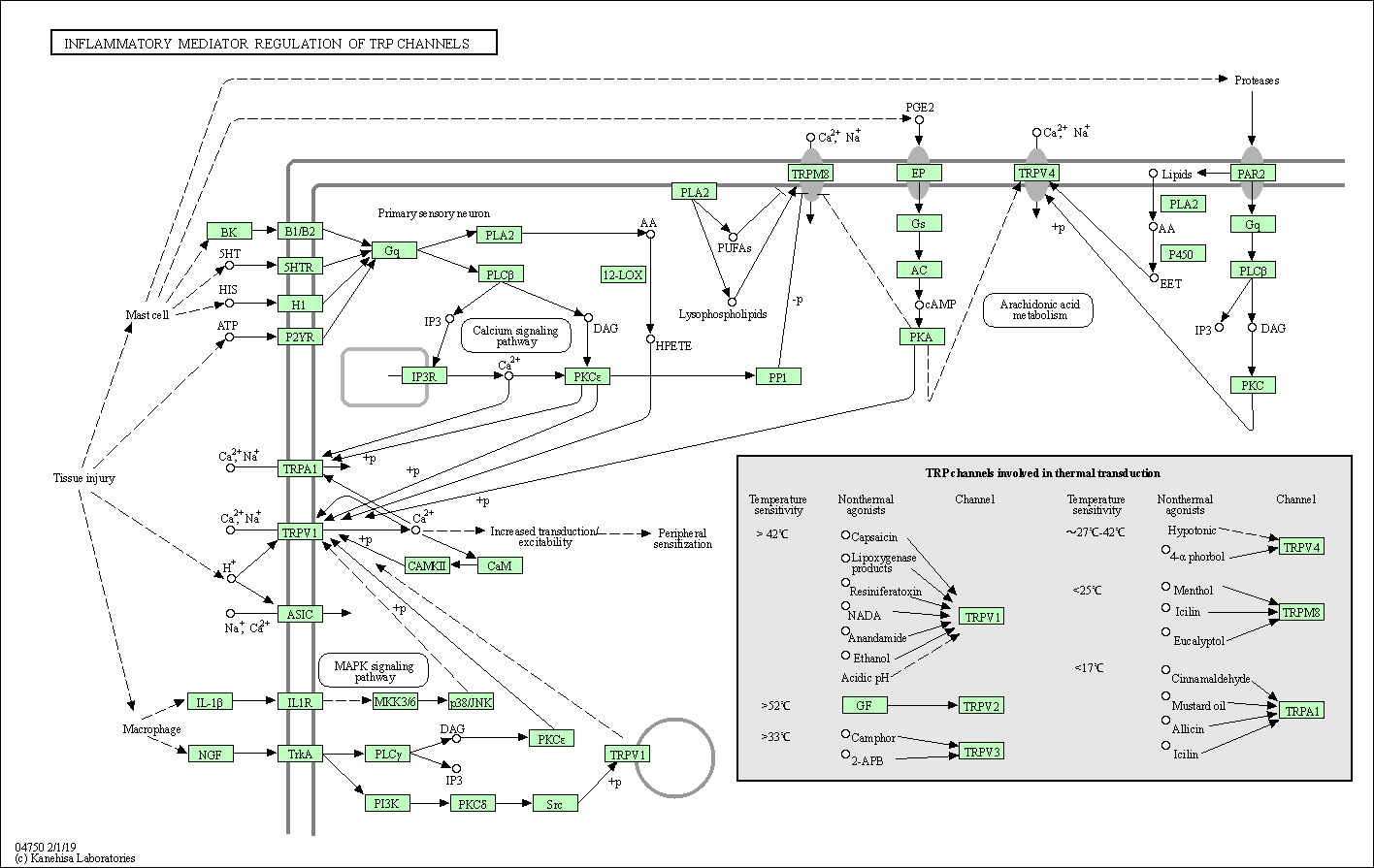
|
|||||||
| cGMP-PKG signaling pathway | hsa04022 |
Pathway Map 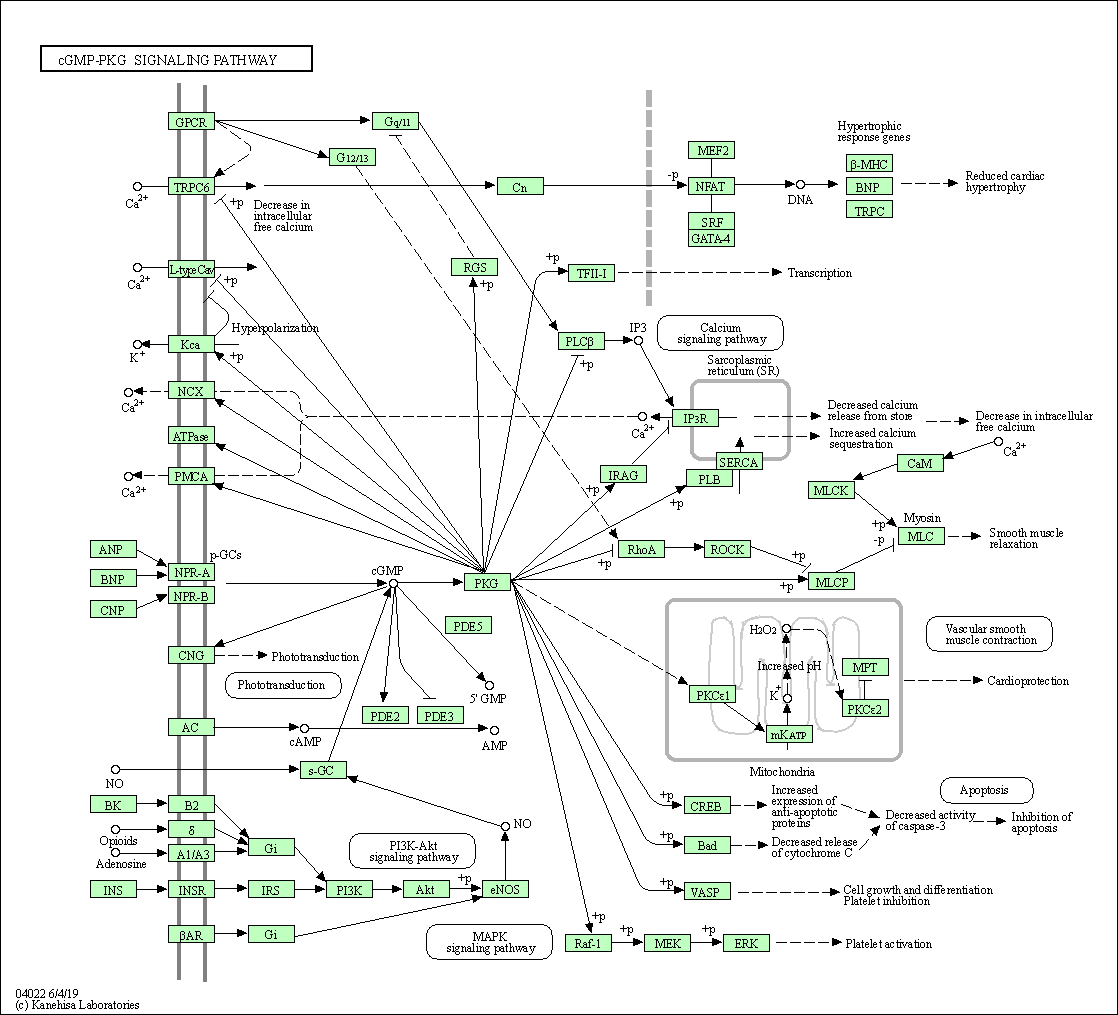
|
|||||||
| cAMP signaling pathway | hsa04024 |
Pathway Map 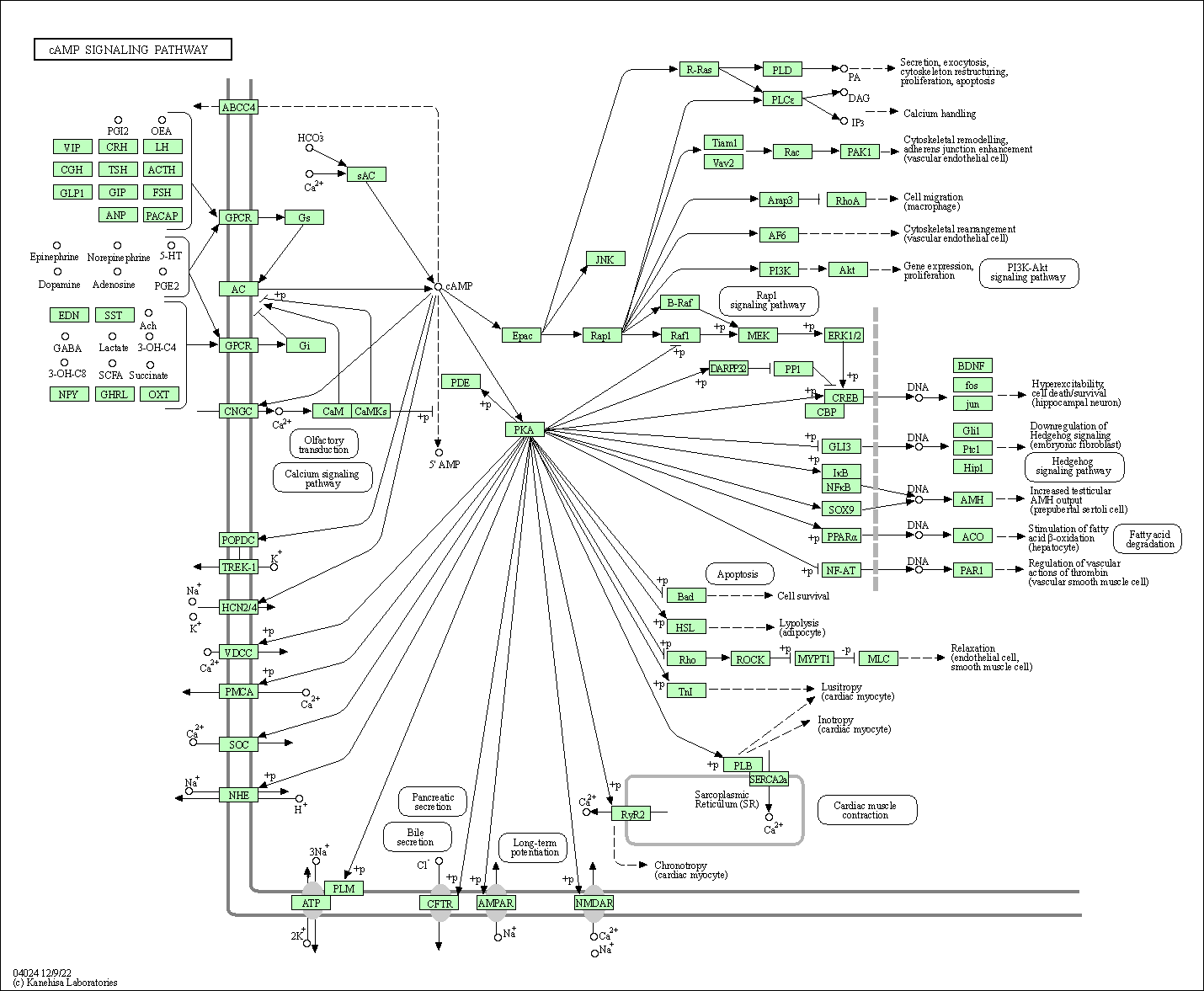
|
|||||||
| Hippo signaling pathway | hsa04390 |
Pathway Map 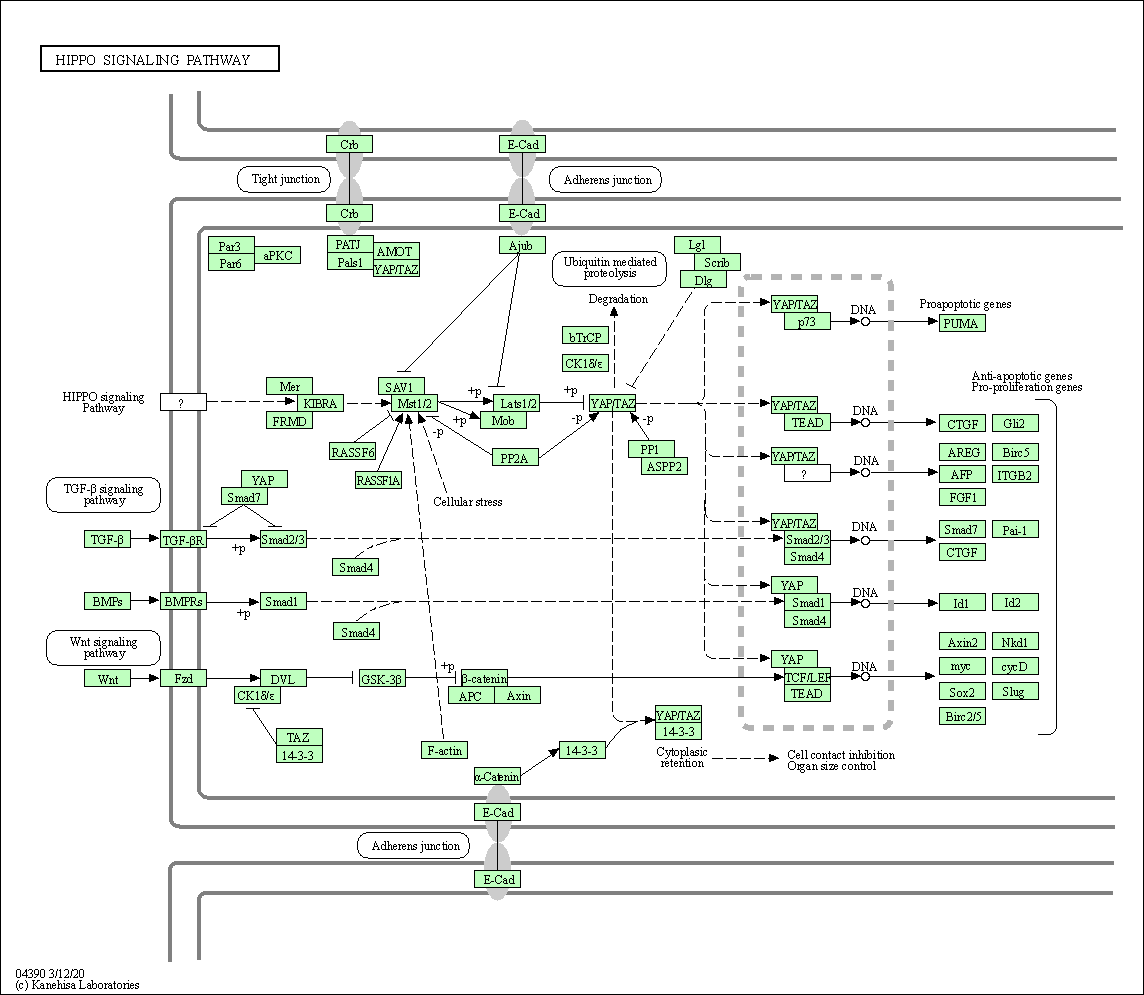
|
|||||||
| Amphetamine addiction | hsa05031 |
Pathway Map 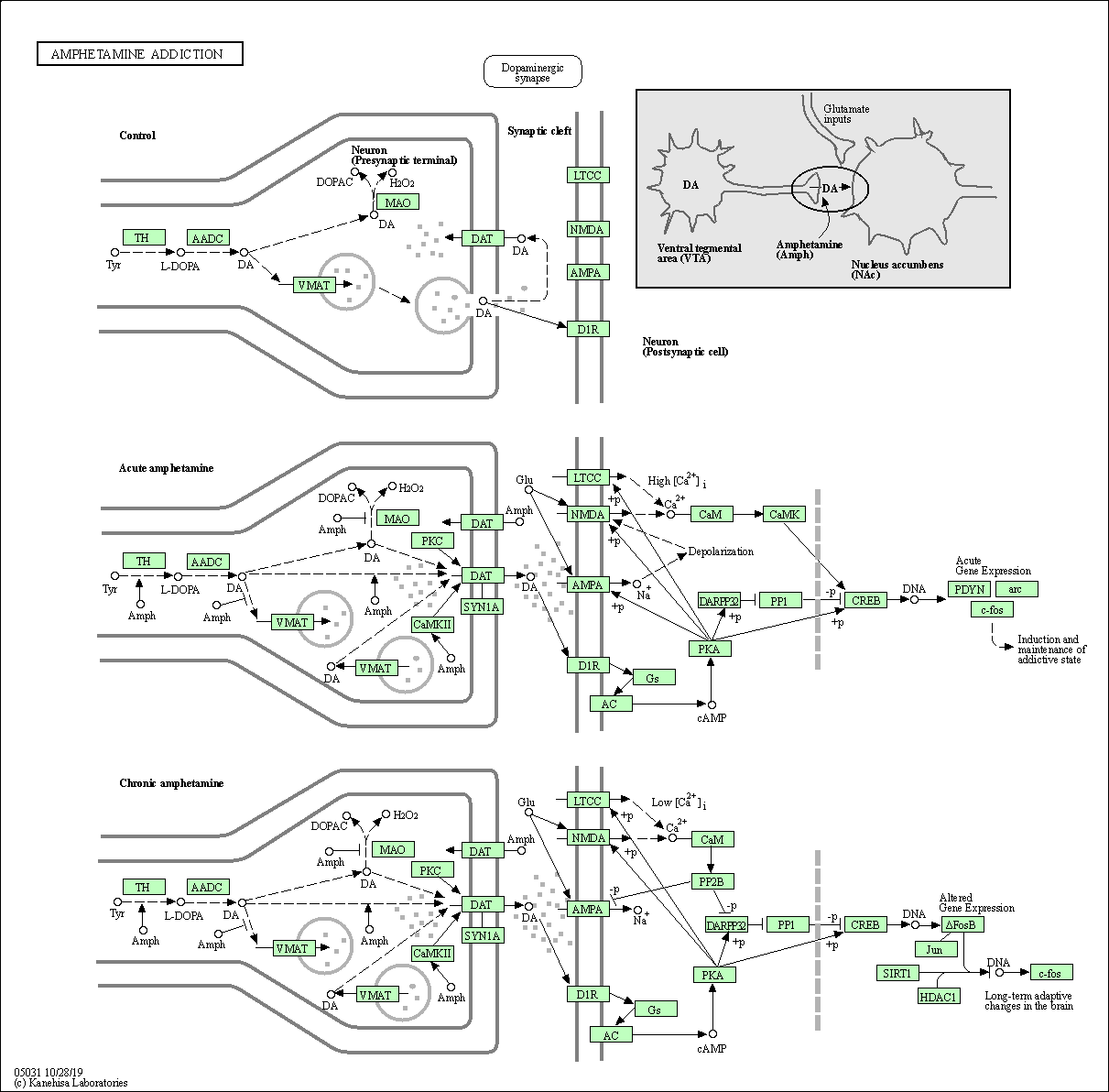
|
|||||||
| Alcoholism | hsa05034 |
Pathway Map 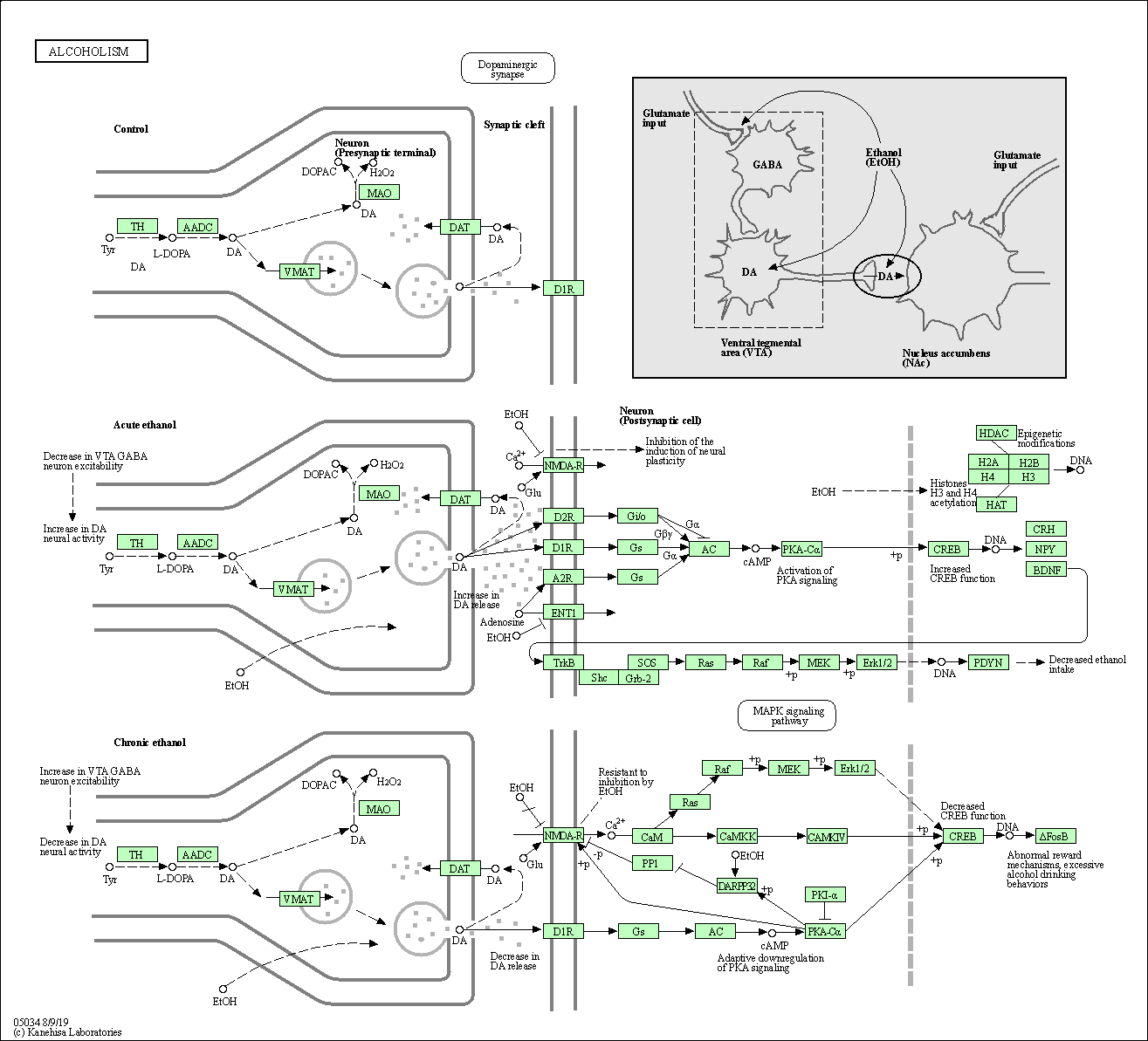
|
|||||||
| mRNA surveillance pathway | hsa03015 |
Pathway Map 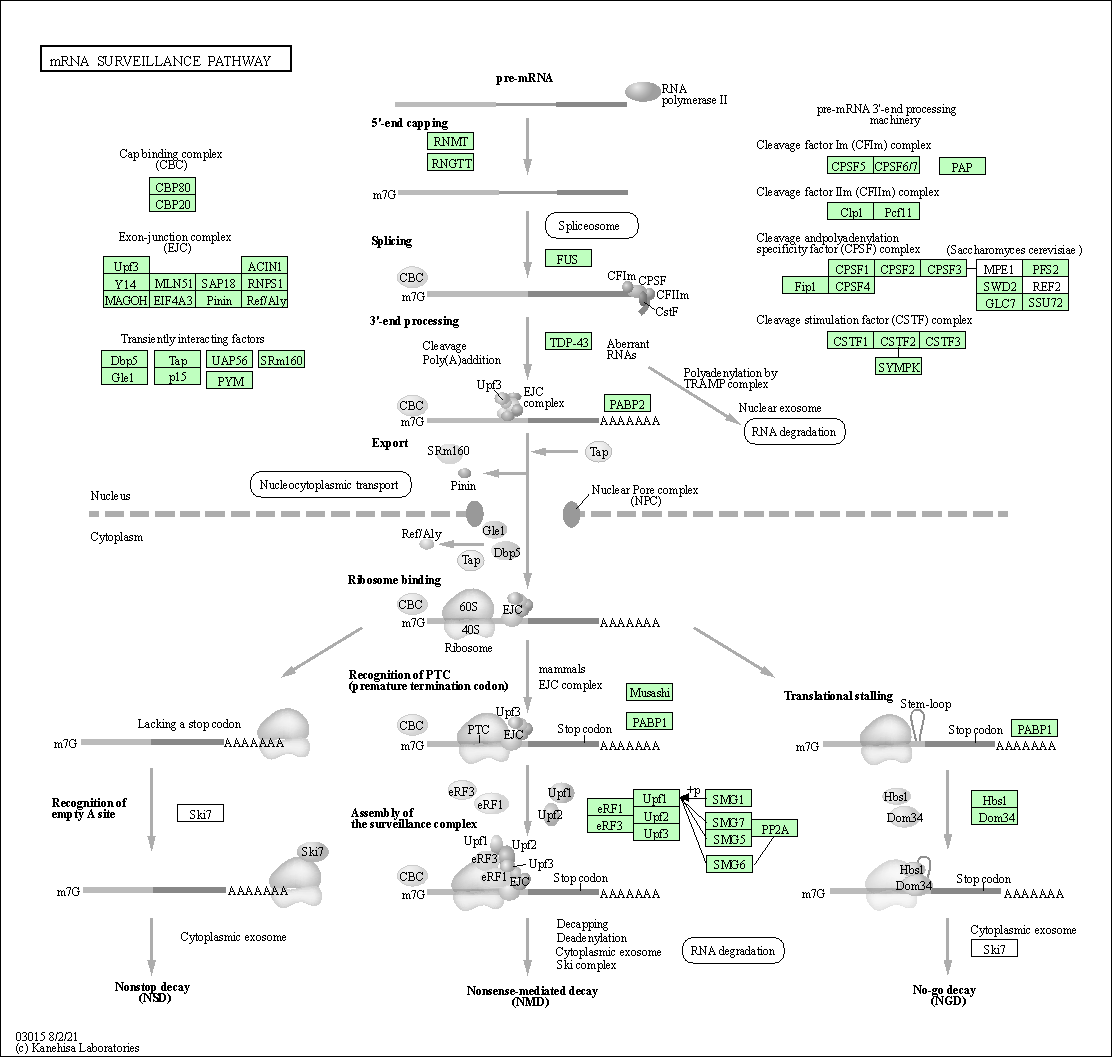
|
|||||||
| 3D Structure |
|
||||||||
Full List of Virus RNA Interacting with This Protien
| Virus Name | Binding Region | RNA Info | Detection Method | Infection Cell | Cell ID | Cell Originated Tissue | Infection Time | Interaction Score 1 | Interaction Score 2 |
|---|---|---|---|---|---|---|---|---|---|
| Encephalomyocarditis virus | 5'UTR - 3'UTR | RNA Info | Comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS) | HuH-7.5 Cells (Human hepatocellular carcinoma cell) | . | Liver | . | Z-score = -5.836536365 | FZR = 3.23514E-06 |
| Influenza A virus | 5'UTR - 3'UTR | RNA Info | Comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS) | HuH-7.5 Cells (Human hepatocellular carcinoma cell) | . | Liver | . | Z-score = 0.274148479 | FZR = 1 |
| Human coronavirus OC43 (strain hCov-OC43/VR-759 Quebec/2019) | Not Specified Virus Region | RNA Info | liquid chromatography with tandem mass spectrometry (LC-MS/MS) | HCT-8 cells (Human ileocaecal adenocarcinoma cell) | . | Ileocecum | 12h; 24; 36h; 48h | p_value < 0.05; FDR < 10% | . |
| Severe acute respiratory syndrome coronavirus 2 (strain hCoV-19/Not Specified Virus Strain) | Not Specified Virus Region | RNA Info | RNAantisensepurificationandquantitativemassspectrometry(RAP-MS); Tandemmasstag(TMT)labelling; liquidchromatographycoupledwithtandemmassspectrometry(LC-MS/MS); Westernblot | Huh7 cells (liver carcinoma celll ine) | . | Liver | 24h | log2 fold change = 7.46165E+14 | . |
Protein Sequence Information
|
MADGELNVDSLITRLLEVRGCRPGKIVQMTEAEVRGLCIKSREIFLSQPILLELEAPLKICGDIHGQYTDLLRLFEYGGFPPEANYLFLGDYVDRGKQSLETICLLLAYKIKYPENFFLLRGNHECASINRIYGFYDECKRRFNIKLWKTFTDCFNCLPIAAIVDEKIFCCHGGLSPDLQSMEQIRRIMRPTDVPDTGLLCDLLWSDPDKDVQGWGENDRGVSFTFGADVVSKFLNRHDLDLICRAHQVVEDGYEFFAKRQLVTLFSAPNYCGEFDNAGGMMSVDETLMCSFQILKPSEKKAKYQYGGLNSGRPVTPPRTANPPKKR
Click to Show/Hide
|
