Host Protein General Information
| Protein Name |
6-phosphofructokinase type A
|
Gene Name |
PFKM
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Host Species |
Homo sapiens
|
Uniprot Entry Name |
PFKAM_HUMAN
|
||||||
| Protein Families |
Phosphofructokinase type A (PFKA) family, ATP-dependent PFK group I subfamily, Eukaryotic two domain clade 'E' sub-subfamily
|
||||||||
| EC Number |
2.7.1.11
|
||||||||
| Subcellular Location |
Cytoplasm
|
||||||||
| External Link | |||||||||
| NCBI Gene ID | |||||||||
| Uniprot ID | |||||||||
| Ensembl ID | |||||||||
| HGNC ID | |||||||||
| Related KEGG Pathway | |||||||||
| Central carbon metabolism in cancer | hsa05230 |
Pathway Map 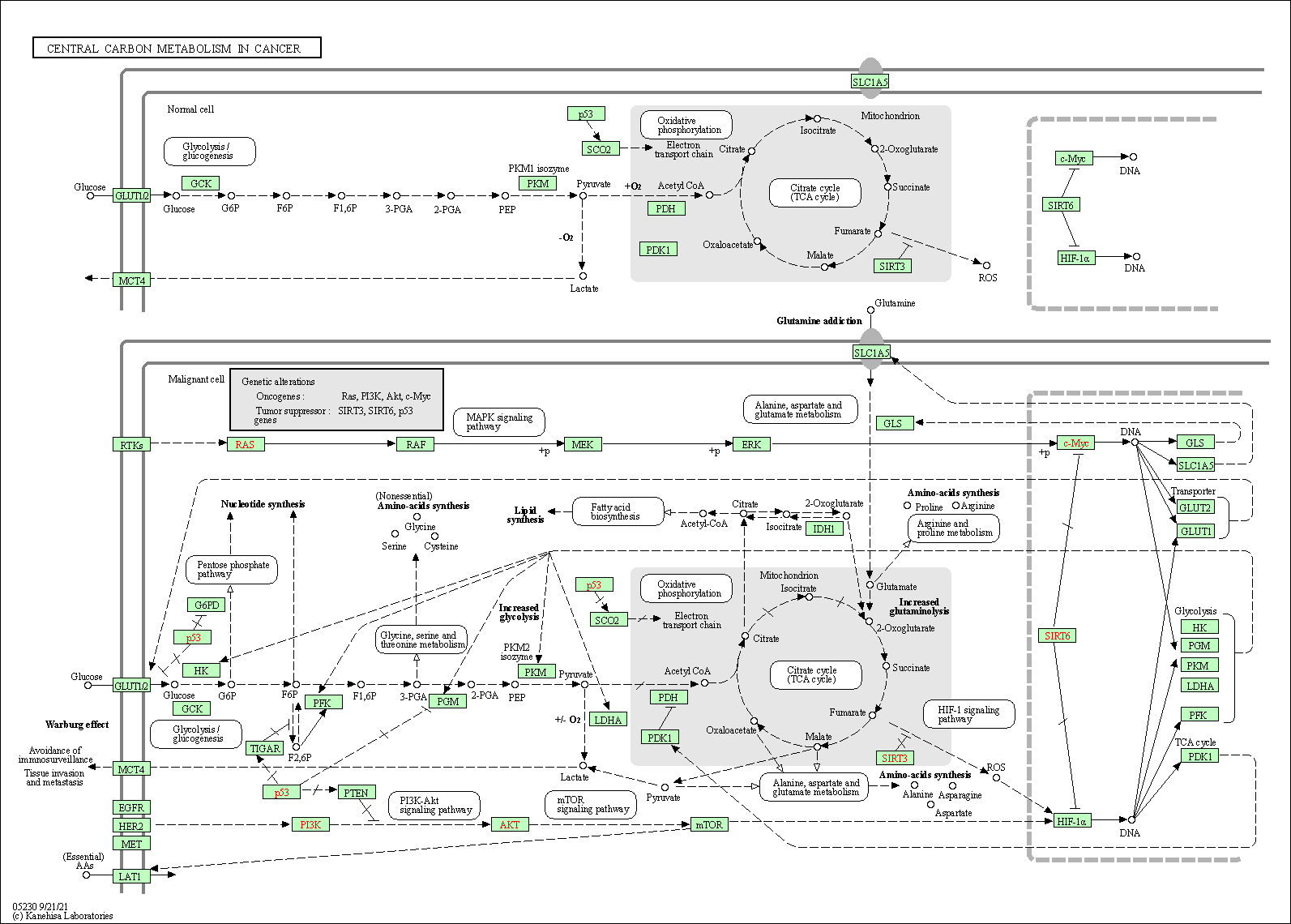
|
|||||||
| Glycolysis / Gluconeogenesis | hsa00010 |
Pathway Map 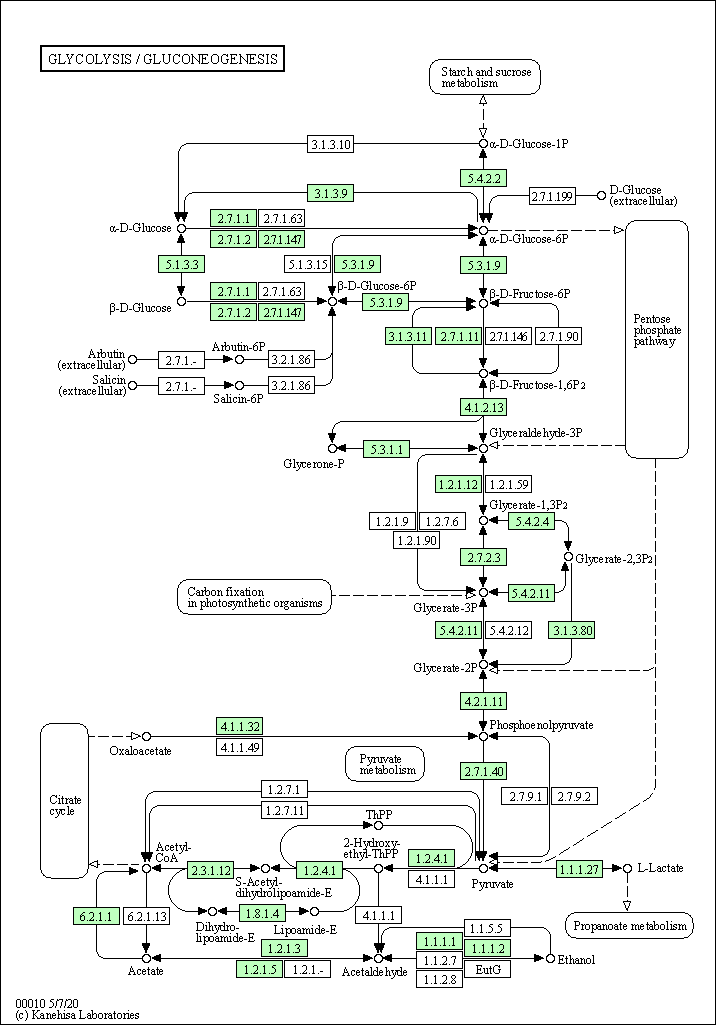
|
|||||||
| Pentose phosphate pathway | hsa00030 |
Pathway Map 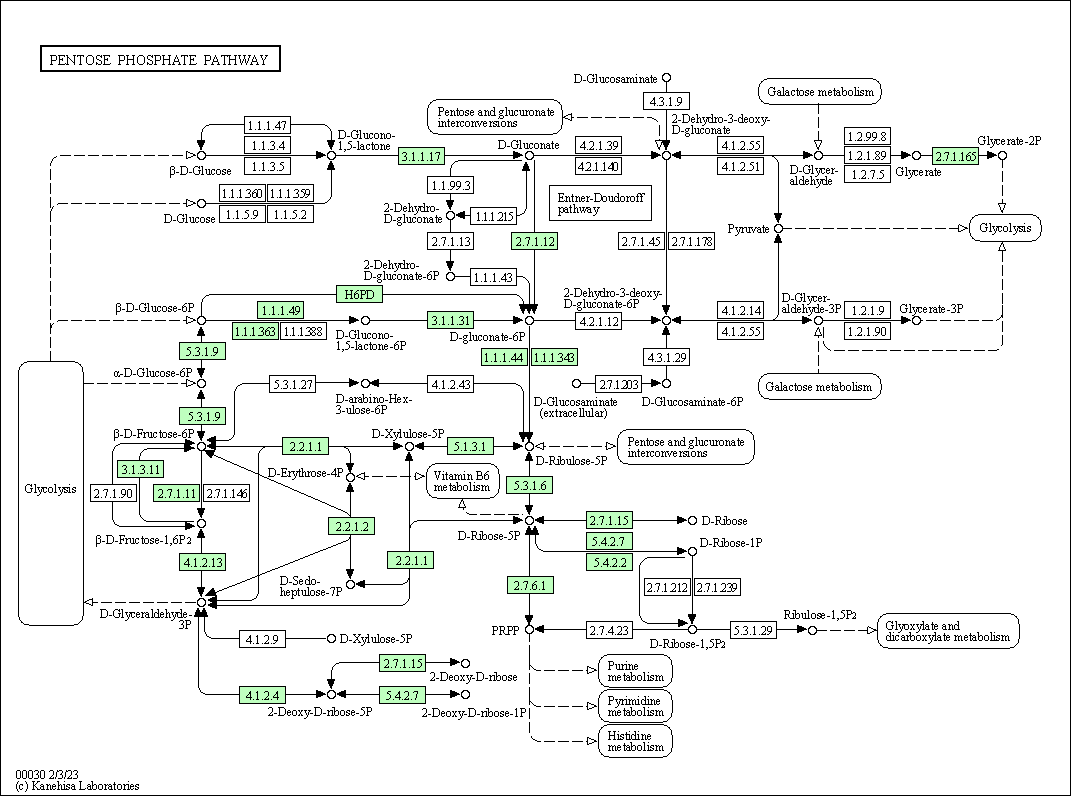
|
|||||||
| Fructose and mannose metabolism | hsa00051 |
Pathway Map 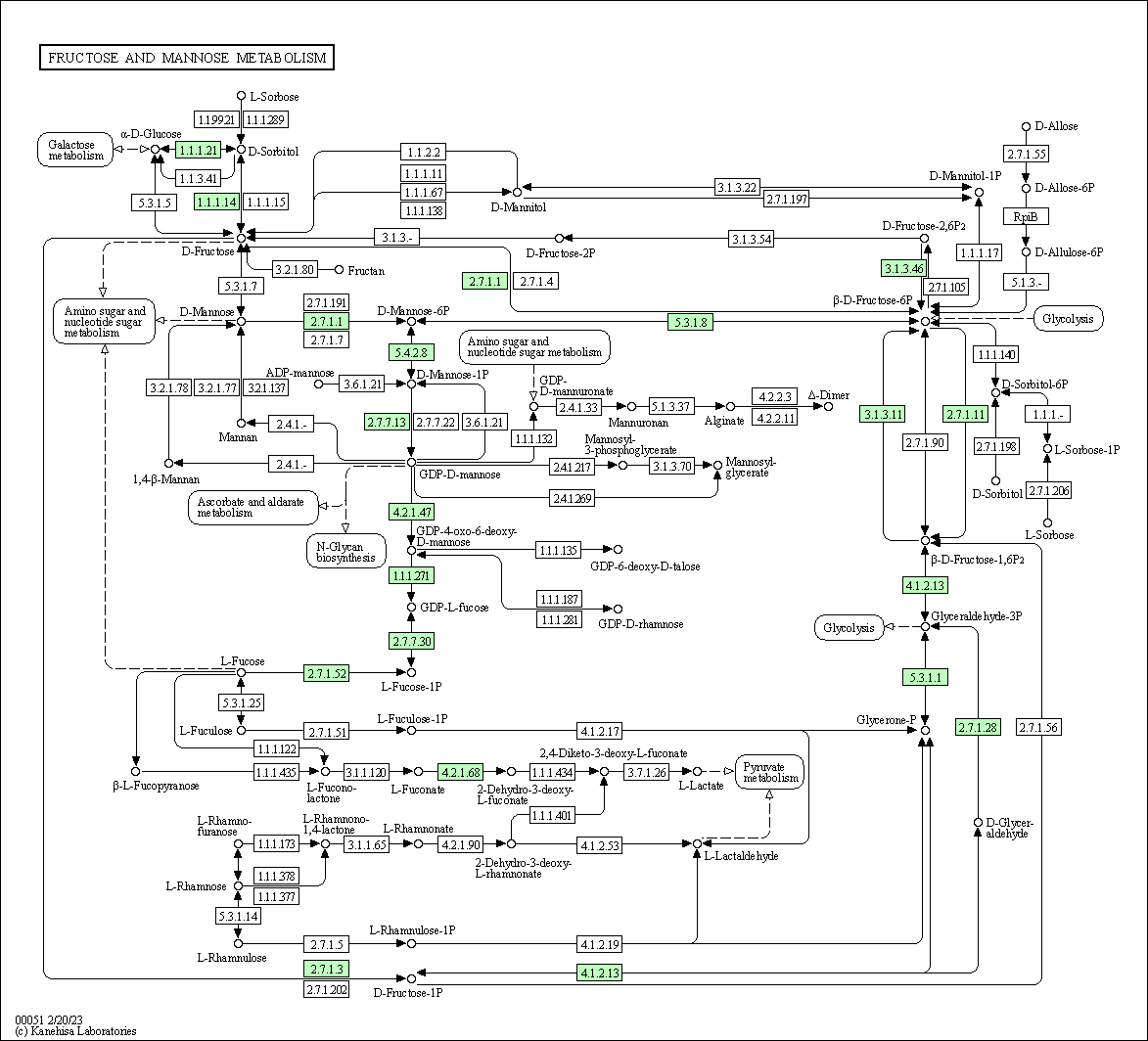
|
|||||||
| Galactose metabolism | hsa00052 |
Pathway Map 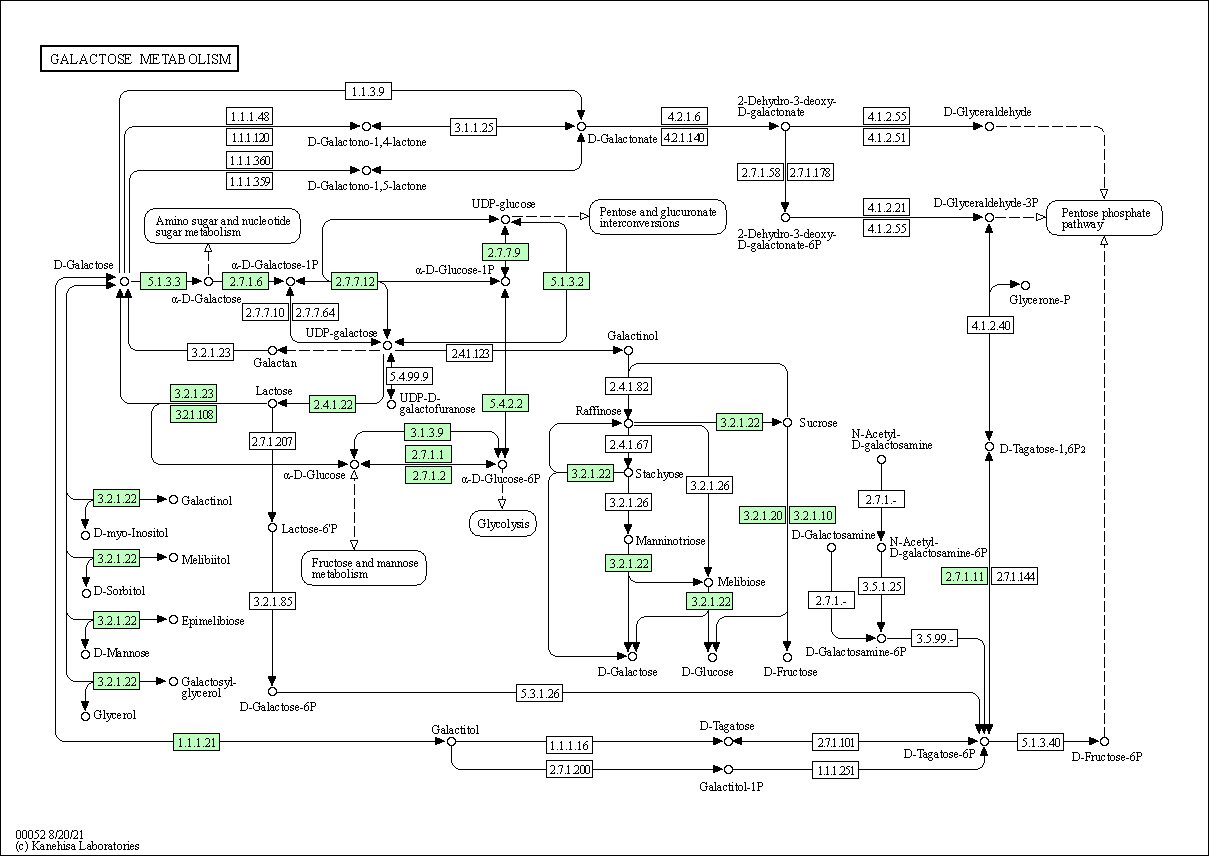
|
|||||||
| Thyroid hormone signaling pathway | hsa04919 |
Pathway Map 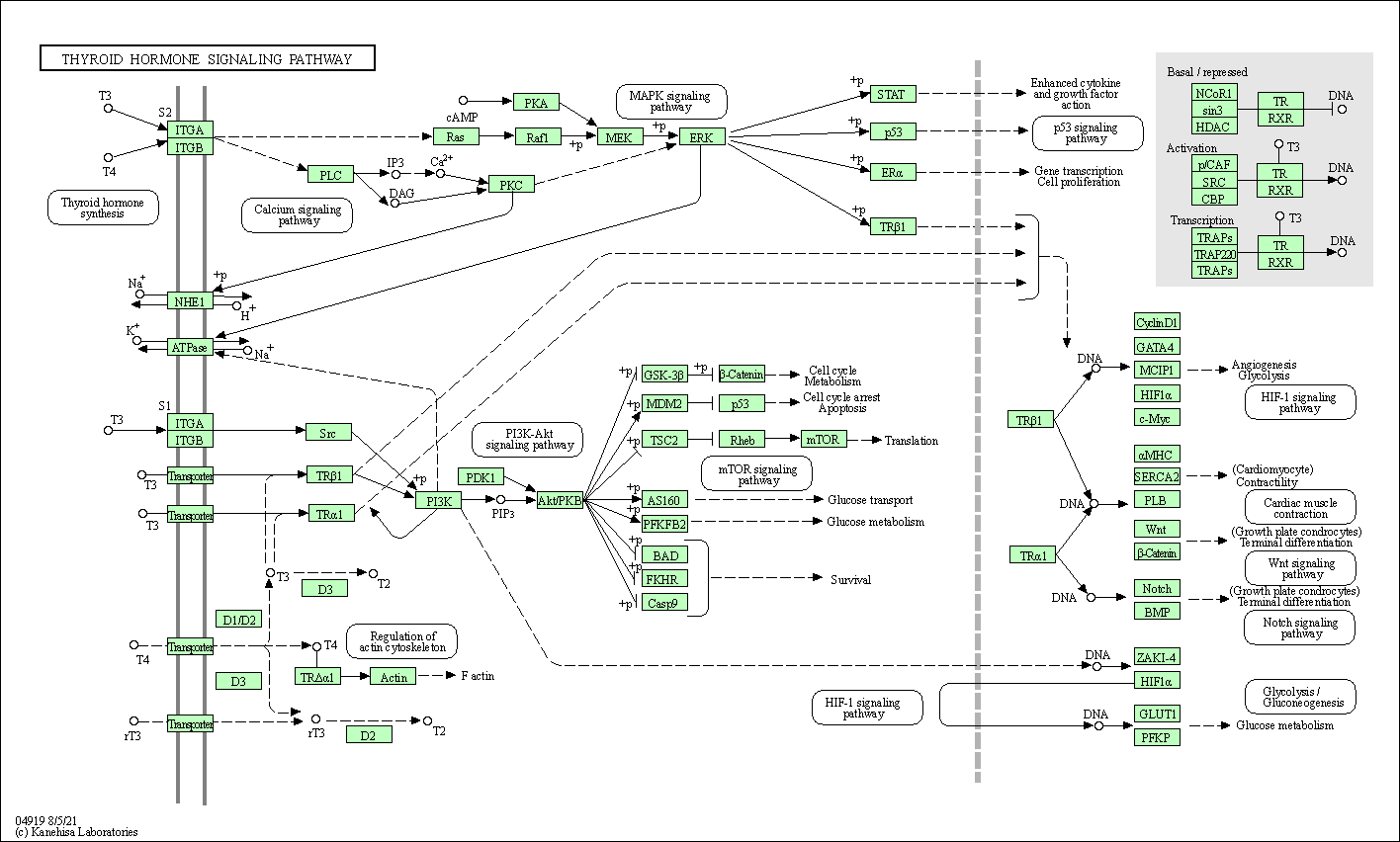
|
|||||||
| Glucagon signaling pathway | hsa04922 |
Pathway Map 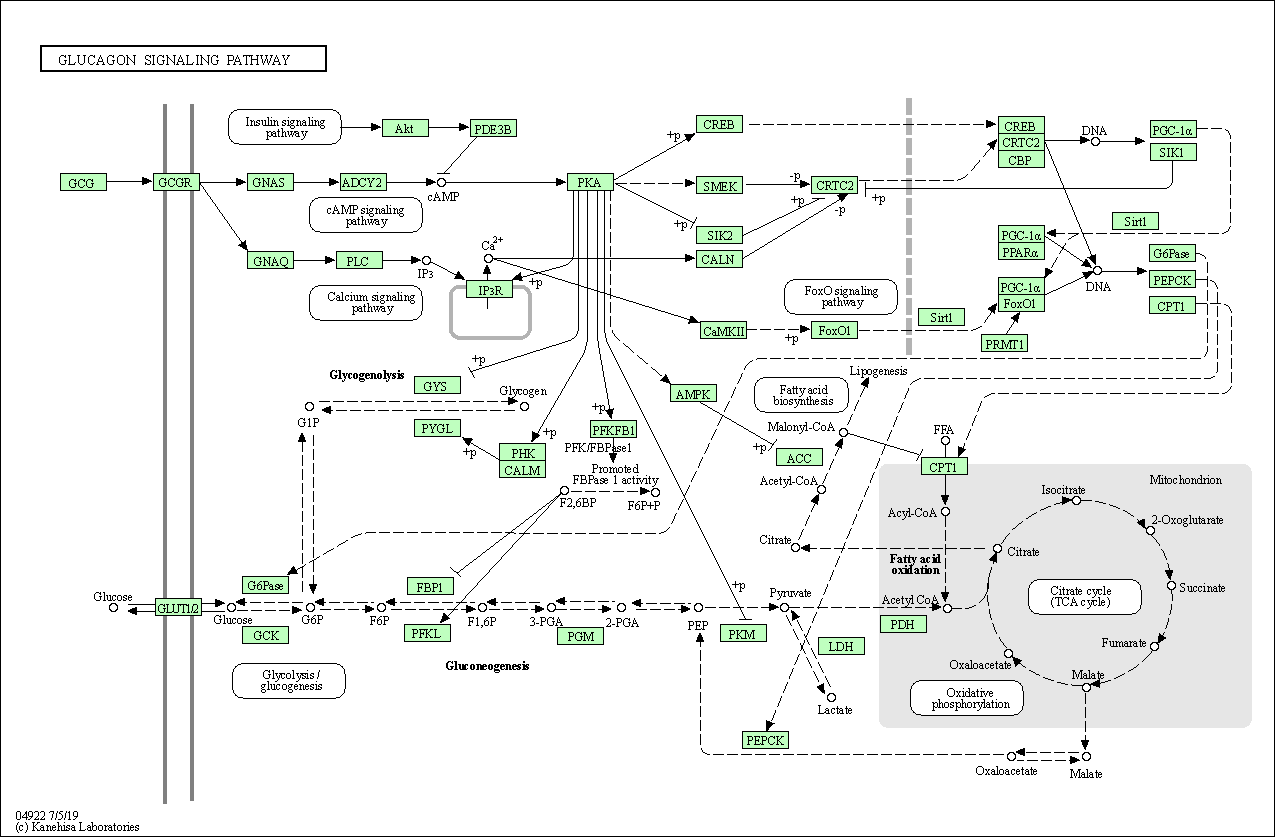
|
|||||||
| RNA degradation | hsa03018 |
Pathway Map 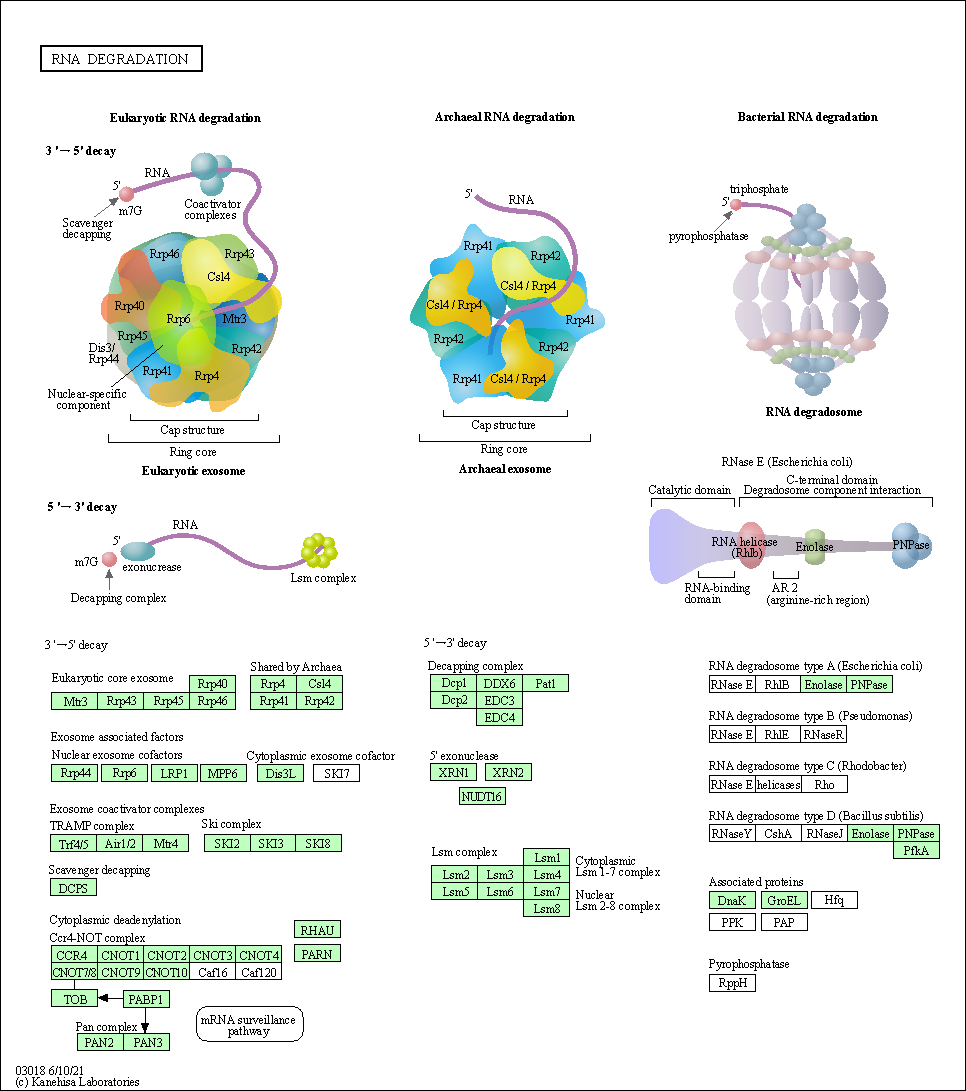
|
|||||||
| Metabolic pathways | hsa01100 |
Pathway Map 
|
|||||||
| Carbon metabolism | hsa01200 |
Pathway Map 
|
|||||||
| Biosynthesis of amino acids | hsa01230 |
Pathway Map 
|
|||||||
| HIF-1 signaling pathway | hsa04066 |
Pathway Map 
|
|||||||
| AMPK signaling pathway | hsa04152 |
Pathway Map 
|
|||||||
| 3D Structure |
|
||||||||
Full List of Virus RNA Interacting with This Protien
| Virus Name | Binding Region | RNA Info | Detection Method | Infection Cell | Cell ID | Cell Originated Tissue | Infection Time | Interaction Score 1 | Interaction Score 2 |
|---|---|---|---|---|---|---|---|---|---|
| Encephalomyocarditis virus | 5'UTR - 3'UTR | RNA Info | Comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS) | HuH-7.5 Cells (Human hepatocellular carcinoma cell) | . | Liver | . | Z-score = 0.934849594 | FZR = 1 |
| Influenza A virus | 5'UTR - 3'UTR | RNA Info | Comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS) | HuH-7.5 Cells (Human hepatocellular carcinoma cell) | . | Liver | . | Z-score = 4.403268767 | FZR = 0.009906106 |
| Chikungunya virus (strain 181/25) | 5'UTR - 3'UTR | RNA Info | VIRal Cross-Linking And Solid-phase Purification (VIR-CLASP) | BHK-21 Cells (Small hamster kidney fibroblast) | . | Kidney | 3 h | . | . |
| Chikungunya virus (strain 181/25) | 5'UTR - 3'UTR | RNA Info | VIRal Cross-Linking And Solid-phase Purification (VIR-CLASP) | BHK-21 Cells (Small hamster kidney fibroblast) | . | Kidney | 1 h | . | . |
| Chikungunya virus (strain 181/25) | 5'UTR - 3'UTR | RNA Info | VIRal Cross-Linking And Solid-phase Purification (VIR-CLASP) | BHK-21 Cells (Small hamster kidney fibroblast) | . | Kidney | 0.2 h | . | . |
Protein Sequence Information
|
MTHEEHHAAKTLGIGKAIAVLTSGGDAQGMNAAVRAVVRVGIFTGARVFFVHEGYQGLVDGGDHIKEATWESVSMMLQLGGTVIGSARCKDFREREGRLRAAYNLVKRGITNLCVIGGDGSLTGADTFRSEWSDLLSDLQKAGKITDEEATKSSYLNIVGLVGSIDNDFCGTDMTIGTDSALHRIMEIVDAITTTAQSHQRTFVLEVMGRHCGYLALVTSLSCGADWVFIPECPPDDDWEEHLCRRLSETRTRGSRLNIIIVAEGAIDKNGKPITSEDIKNLVVKRLGYDTRVTVLGHVQRGGTPSAFDRILGSRMGVEAVMALLEGTPDTPACVVSLSGNQAVRLPLMECVQVTKDVTKAMDEKKFDEALKLRGRSFMNNWEVYKLLAHVRPPVSKSGSHTVAVMNVGAPAAGMNAAVRSTVRIGLIQGNRVLVVHDGFEGLAKGQIEEAGWSYVGGWTGQGGSKLGTKRTLPKKSFEQISANITKFNIQGLVIIGGFEAYTGGLELMEGRKQFDELCIPFVVIPATVSNNVPGSDFSVGADTALNTICTTCDRIKQSAAGTKRRVFIIETMGGYCGYLATMAGLAAGADAAYIFEEPFTIRDLQANVEHLVQKMKTTVKRGLVLRNEKCNENYTTDFIFNLYSEEGKGIFDSRKNVLGHMQQGGSPTPFDRNFATKMGAKAMNWMSGKIKESYRNGRIFANTPDSGCVLGMRKRALVFQPVAELKDQTDFEHRIPKEQWWLKLRPILKILAKYEIDLDTSDHAHLEHITRKRSGEAAV
Click to Show/Hide
|
