Host Protein General Information
| Protein Name |
Ribose-phosphate pyrophosphokinase 1
|
Gene Name |
PRPS1
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Host Species |
Homo sapiens
|
Uniprot Entry Name |
PRPS1_HUMAN
|
||||||
| Protein Families |
Ribose-phosphate pyrophosphokinase family
|
||||||||
| EC Number |
2.7.6.1
|
||||||||
| External Link | |||||||||
| NCBI Gene ID | |||||||||
| Uniprot ID | |||||||||
| Ensembl ID | |||||||||
| HGNC ID | |||||||||
| Related KEGG Pathway | |||||||||
| Pentose phosphate pathway | hsa00030 |
Pathway Map 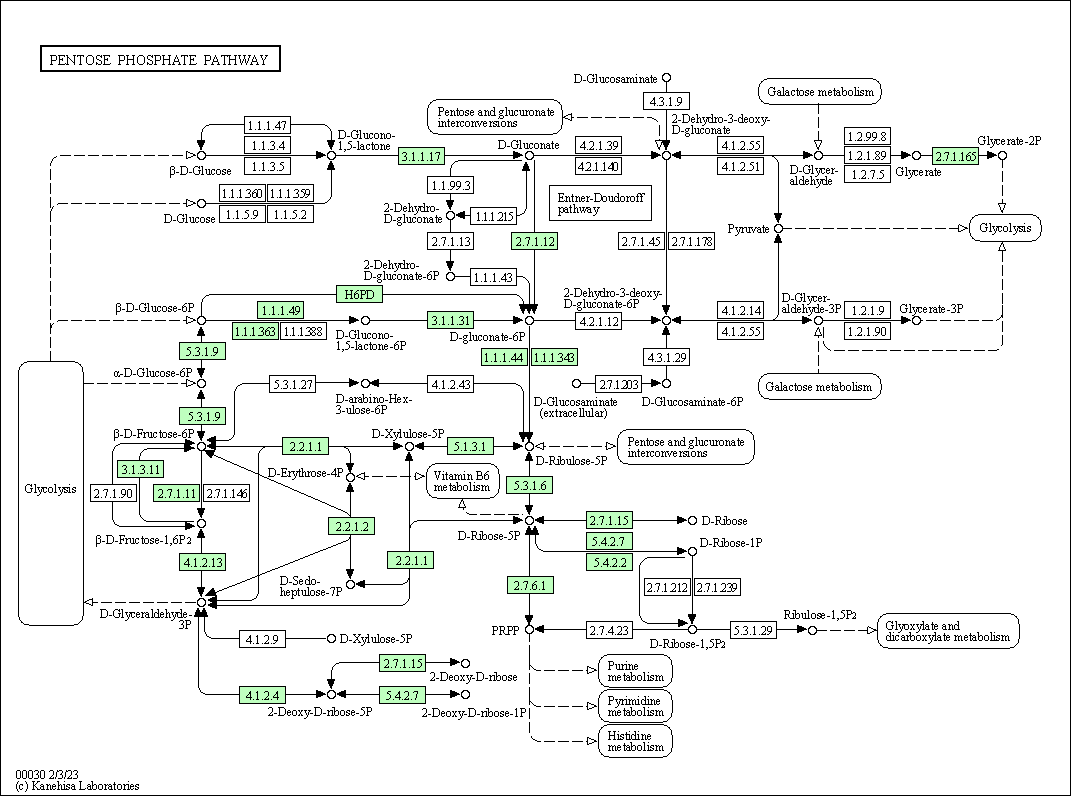
|
|||||||
| Metabolic pathways | hsa01100 |
Pathway Map 
|
|||||||
| Carbon metabolism | hsa01200 |
Pathway Map 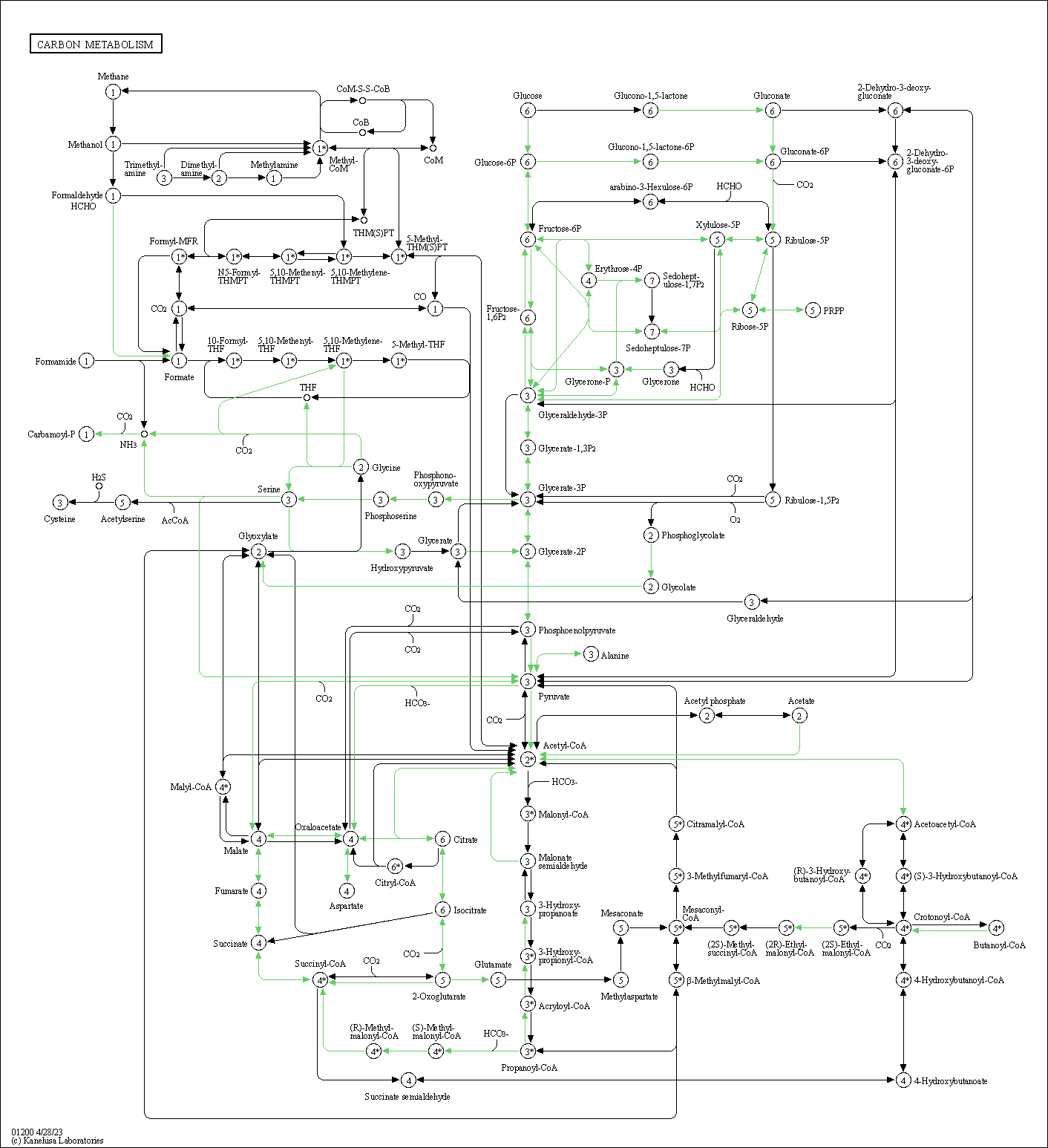
|
|||||||
| Biosynthesis of amino acids | hsa01230 |
Pathway Map 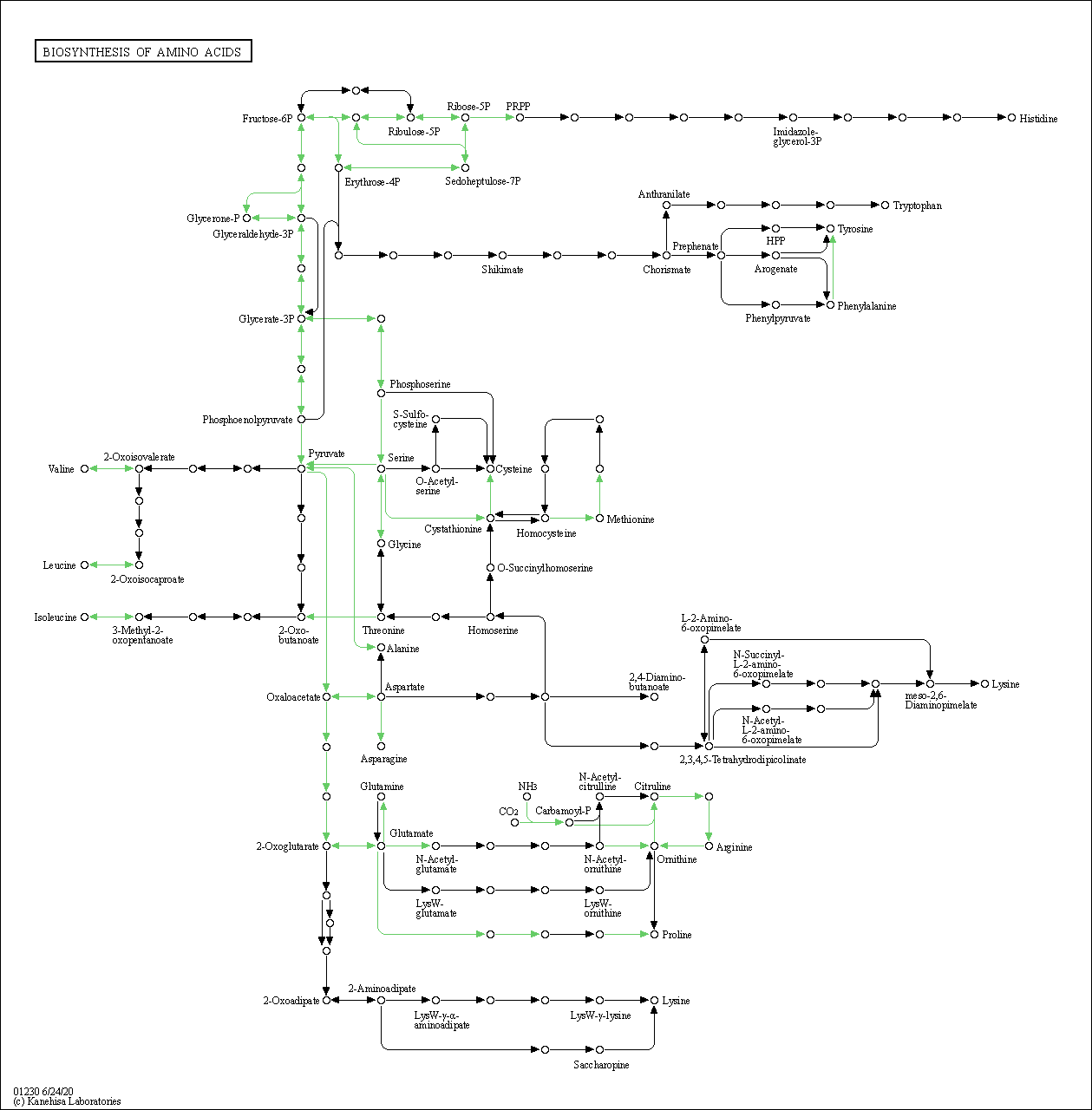
|
|||||||
| Purine metabolism | hsa00230 |
Pathway Map 
|
|||||||
| 3D Structure |
|
||||||||
Full List of Virus RNA Interacting with This Protien
| Virus Name | Binding Region | RNA Info | Detection Method | Infection Cell | Cell ID | Cell Originated Tissue | Infection Time | Interaction Score 1 | Interaction Score 2 |
|---|---|---|---|---|---|---|---|---|---|
| Encephalomyocarditis virus | 5'UTR - 3'UTR | RNA Info | Comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS) | HuH-7.5 Cells (Human hepatocellular carcinoma cell) | . | Liver | . | Z-score = -7.654696378 | FZR = 1.49203E-11 |
| Influenza A virus | 5'UTR - 3'UTR | RNA Info | Comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS) | HuH-7.5 Cells (Human hepatocellular carcinoma cell) | . | Liver | . | Z-score = -0.139377913 | FZR = 1 |
| Dengue virus 2 (strain NGC) | 5'UTR - 3'UTR | RNA Info | VIRal Cross-Linking And Solid-phase Purification (VIR-CLASP) | HuH-7 Cells (Human hepatocellular carcinoma cell) | . | Liver | . | . | . |
Potential Drug(s) that Targets This Protein
| Drug Name | DrunkBank ID | Pubchem ID | TTD ID | REF | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| clofarabine | . | 119182 | D0R5RR | DrugCentral | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| MERCAPTOPURINE | . | 667490 | D09UZO | DGIdb | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| THIOGUANINE | . | 2723601 | D02ZXM | DGIdb | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| vidarabine | . | 21704 | D0NI0C | DrugCentral | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Protein Sequence Information
|
MPNIKIFSGSSHQDLSQKIADRLGLELGKVVTKKFSNQETCVEIGESVRGEDVYIVQSGCGEINDNLMELLIMINACKIASASRVTAVIPCFPYARQDKKDKSRAPISAKLVANMLSVAGADHIITMDLHASQIQGFFDIPVDNLYAEPAVLKWIRENISEWRNCTIVSPDAGGAKRVTSIADRLNVDFALIHKERKKANEVDRMVLVGDVKDRVAILVDDMADTCGTICHAADKLLSAGATRVYAILTHGIFSGPAISRINNACFEAVVVTNTIPQEDKMKHCSKIQVIDISMILAEAIRRTHNGESVSYLFSHVPL
Click to Show/Hide
|
