Host Protein General Information
| Protein Name |
Transcription factor p65
|
Gene Name |
RELA
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Host Species |
Homo sapiens
|
Uniprot Entry Name |
TF65_HUMAN
|
||||||
| Subcellular Location |
Nucleus
|
||||||||
| External Link | |||||||||
| NCBI Gene ID | |||||||||
| Uniprot ID | |||||||||
| Ensembl ID | |||||||||
| HGNC ID | |||||||||
| Related KEGG Pathway | |||||||||
| Chemokine signaling pathway | hsa04062 |
Pathway Map 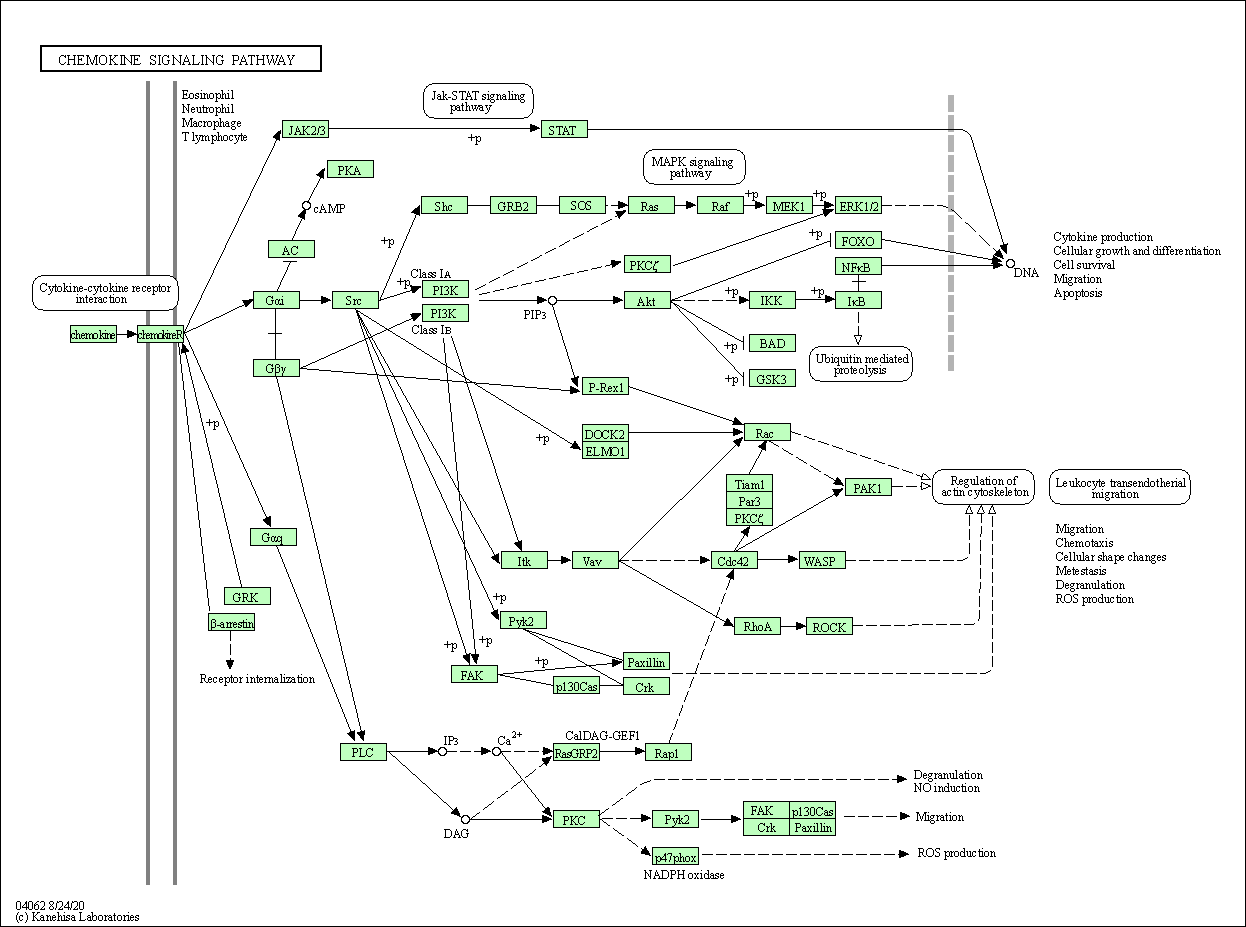
|
|||||||
| Neutrophil extracellular trap formation | hsa04613 |
Pathway Map 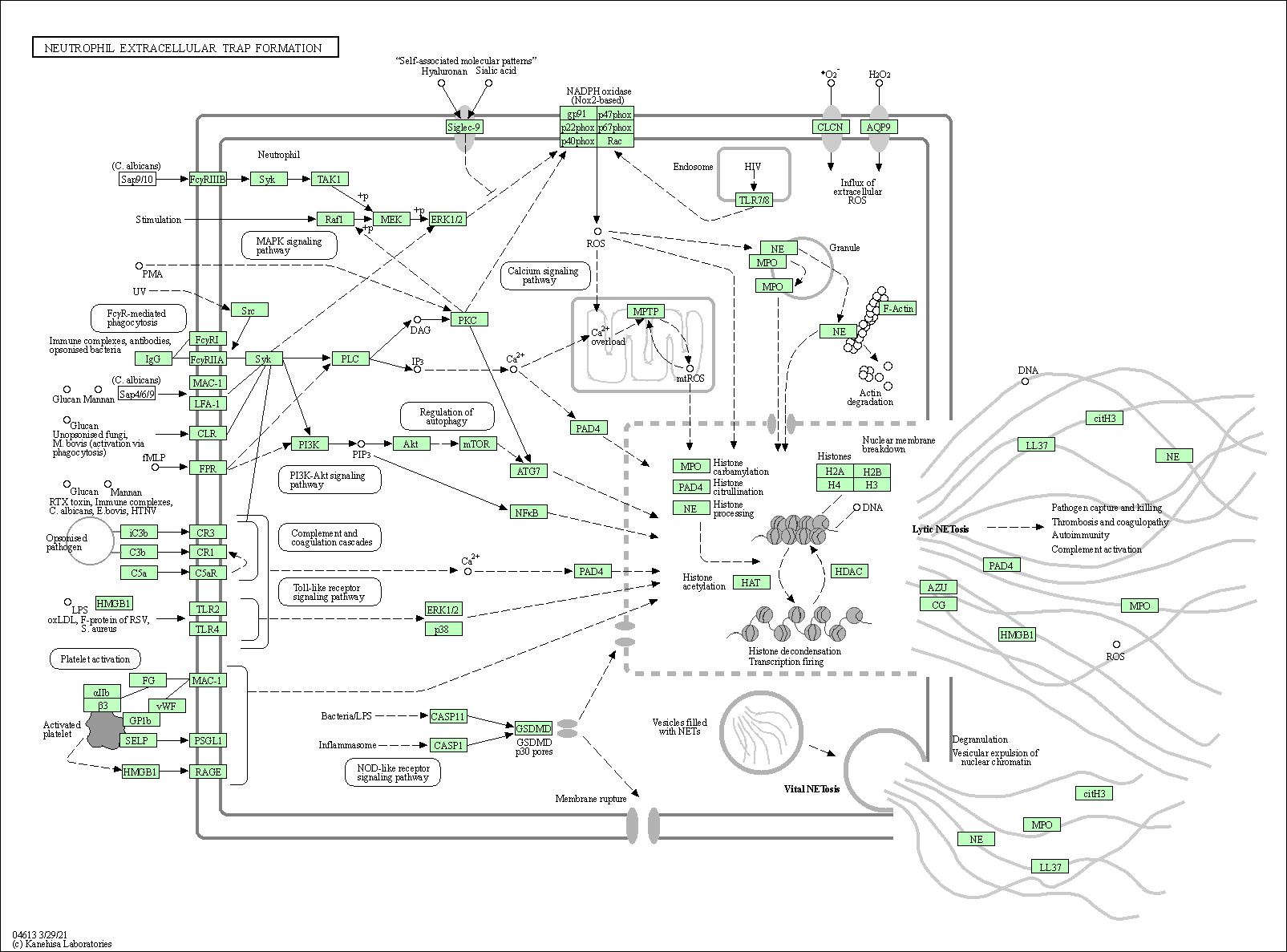
|
|||||||
| Toll-like receptor signaling pathway | hsa04620 |
Pathway Map 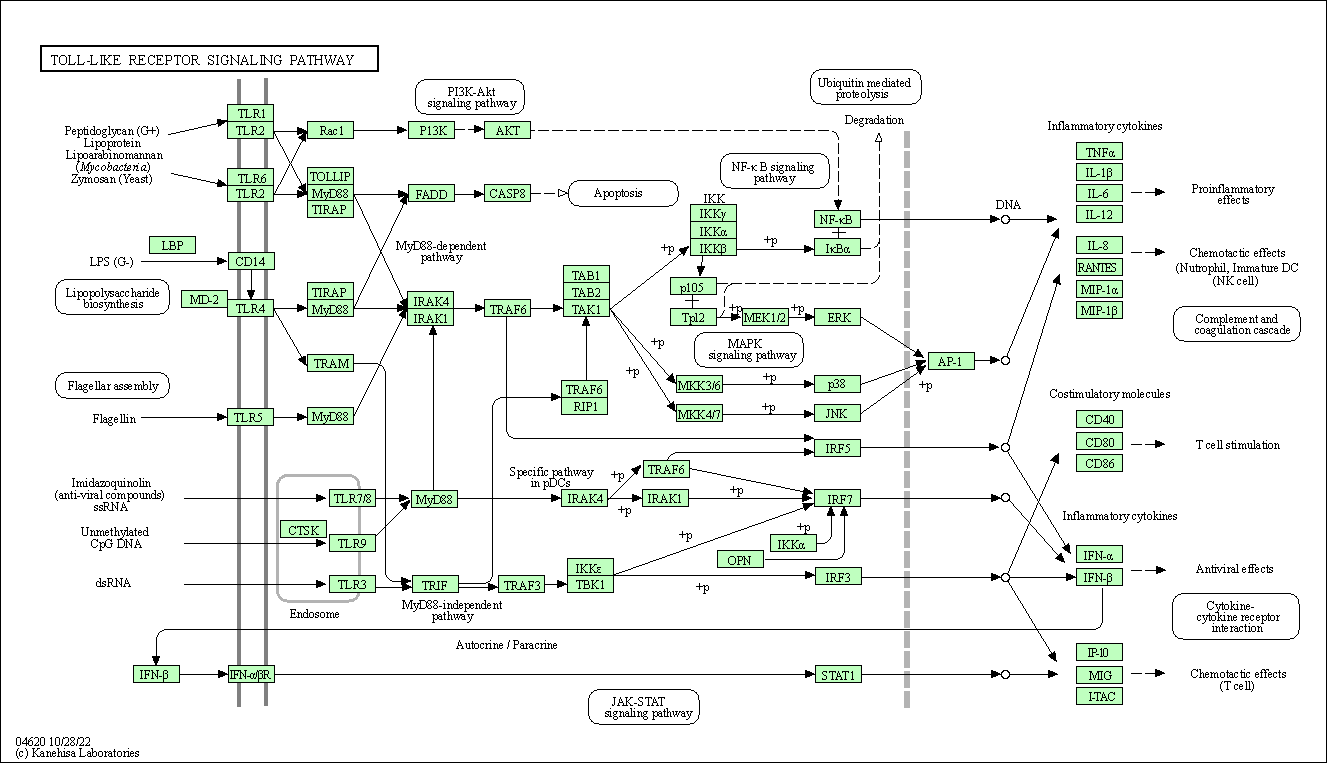
|
|||||||
| NOD-like receptor signaling pathway | hsa04621 |
Pathway Map 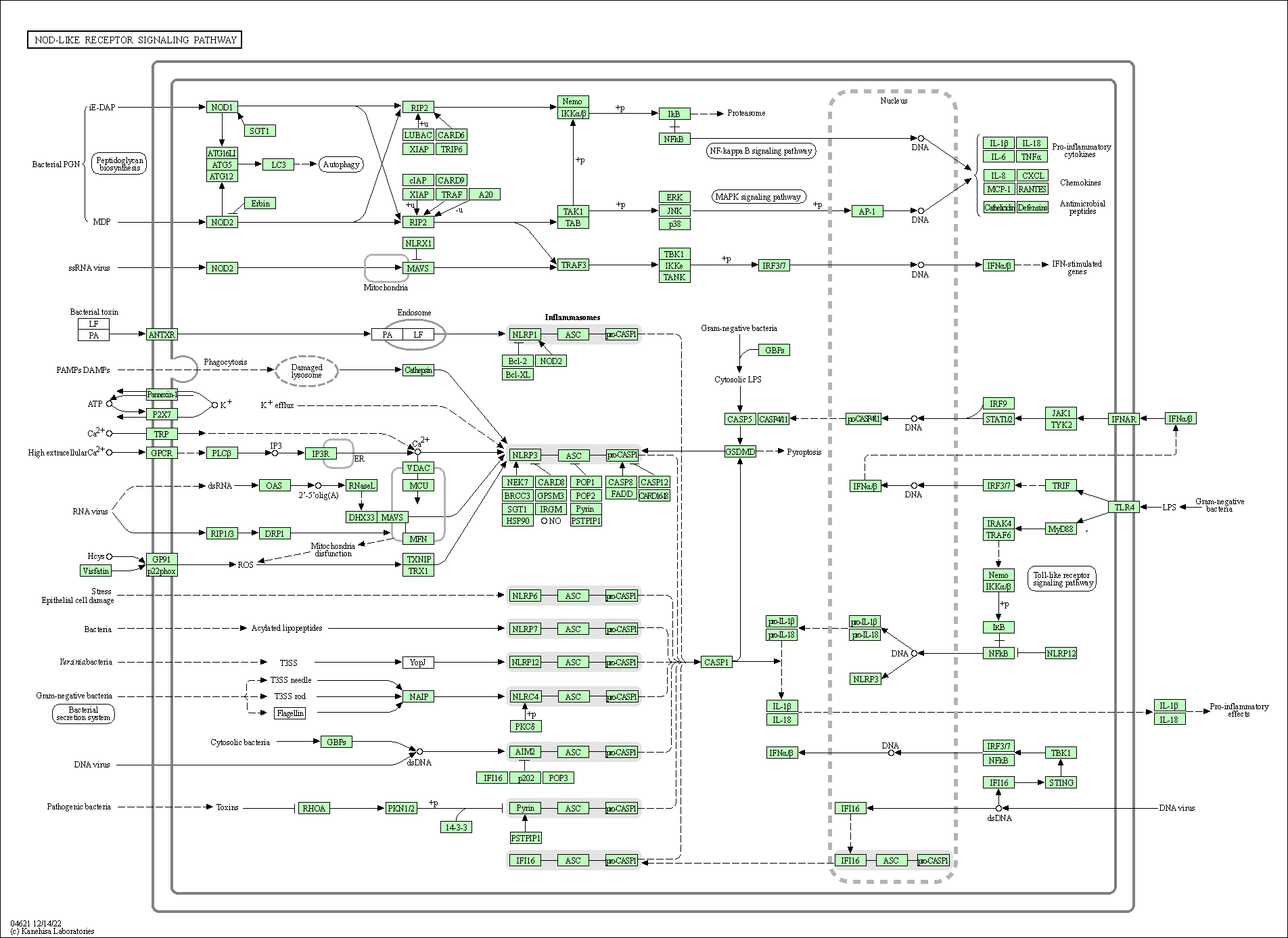
|
|||||||
| RIG-I-like receptor signaling pathway | hsa04622 |
Pathway Map 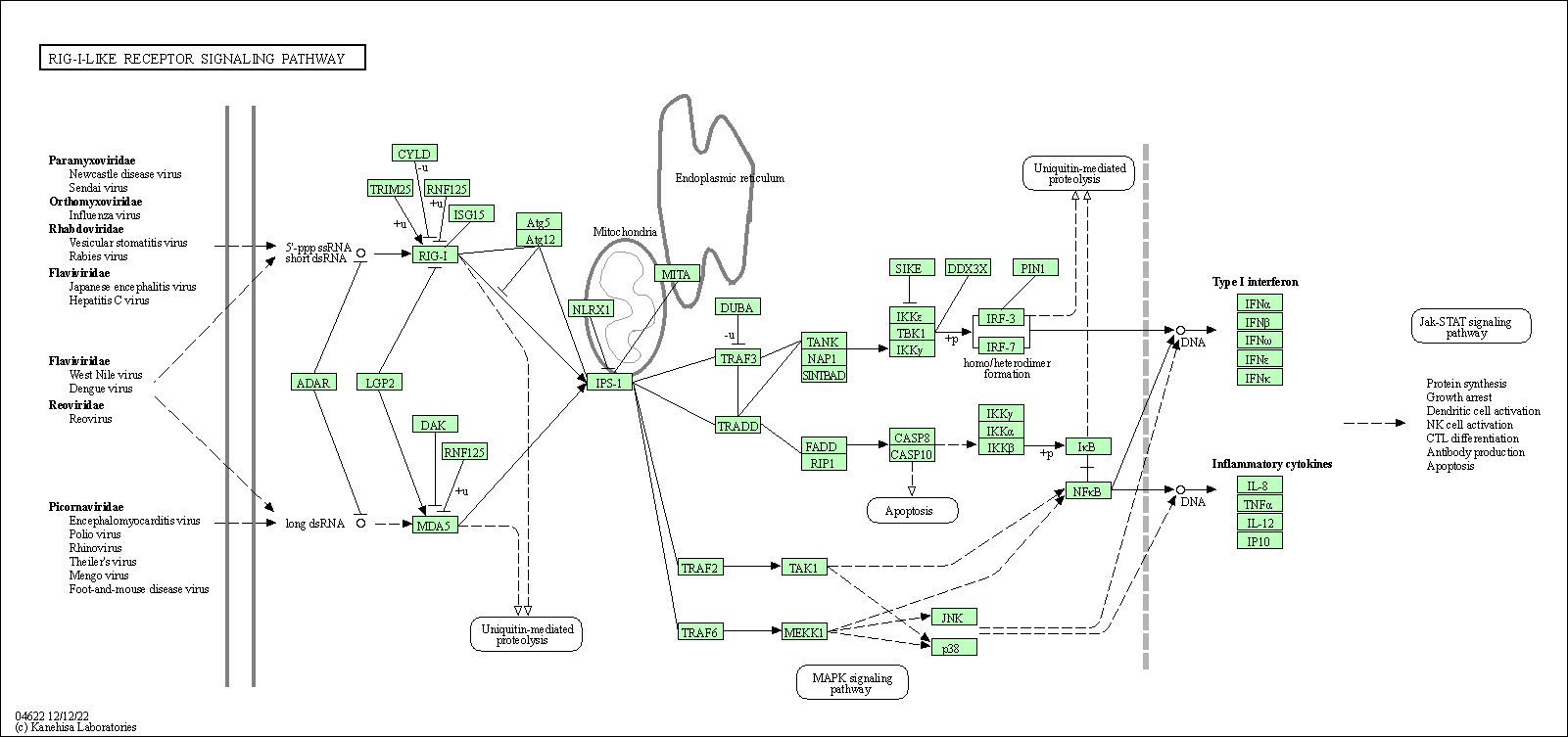
|
|||||||
| Cytosolic DNA-sensing pathway | hsa04623 |
Pathway Map 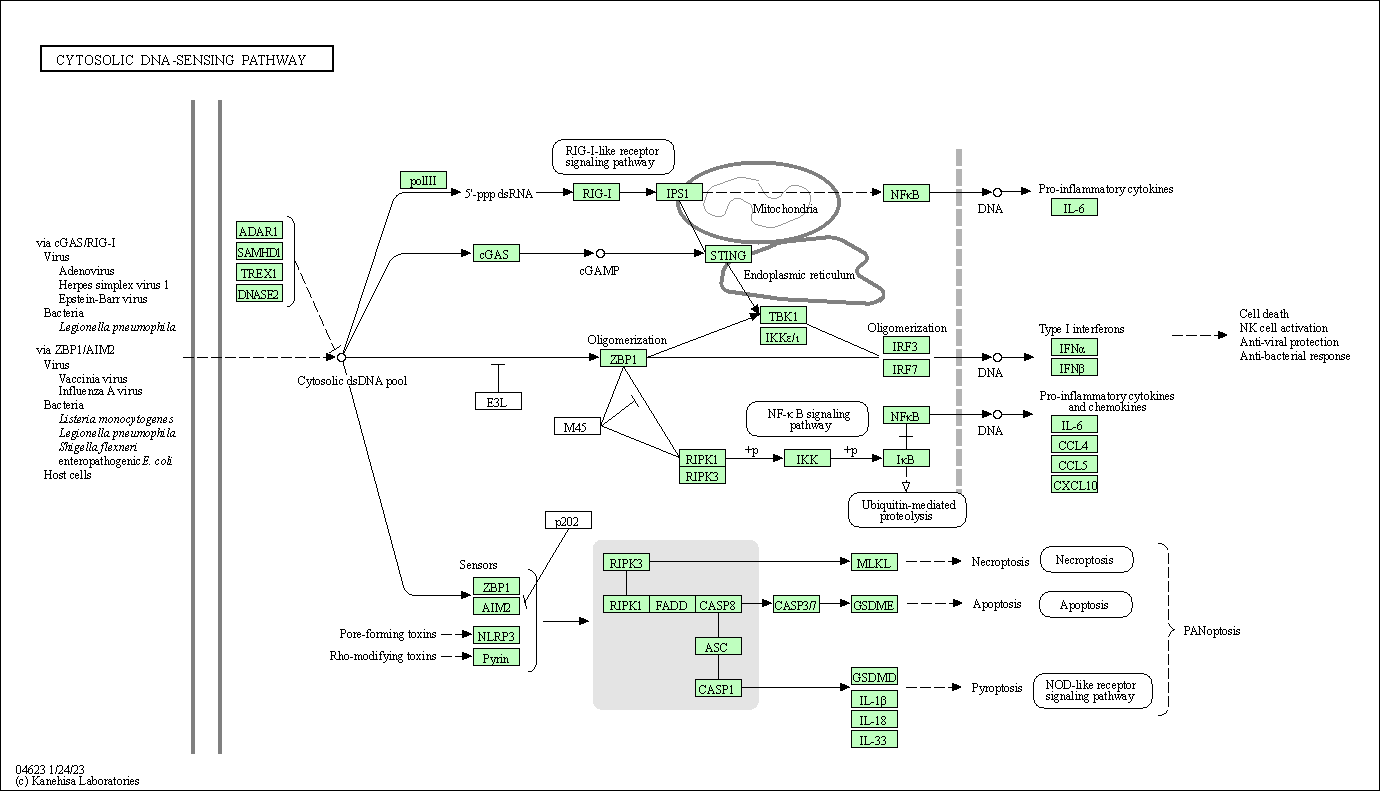
|
|||||||
| C-type lectin receptor signaling pathway | hsa04625 |
Pathway Map 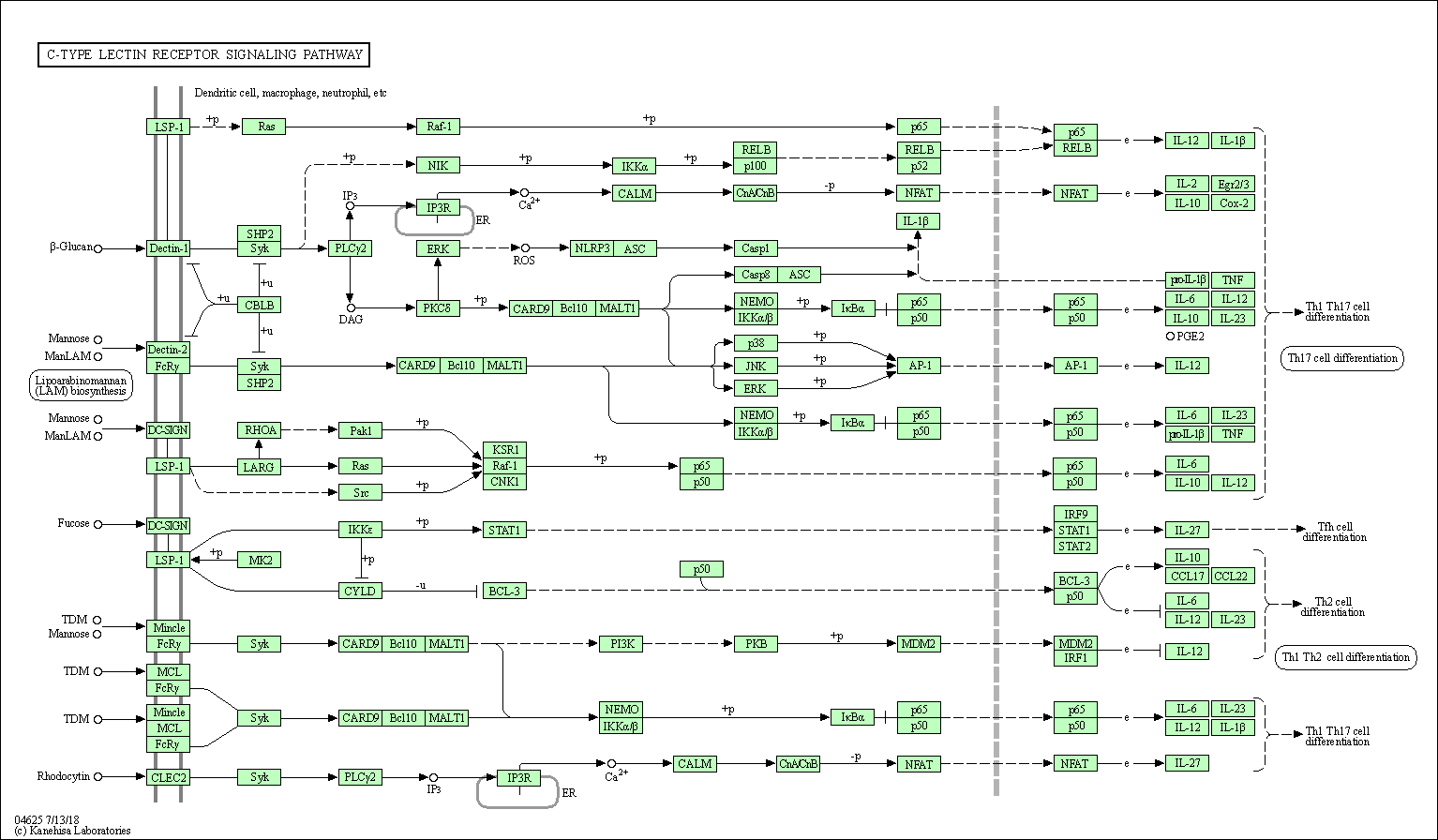
|
|||||||
| IL-17 signaling pathway | hsa04657 |
Pathway Map 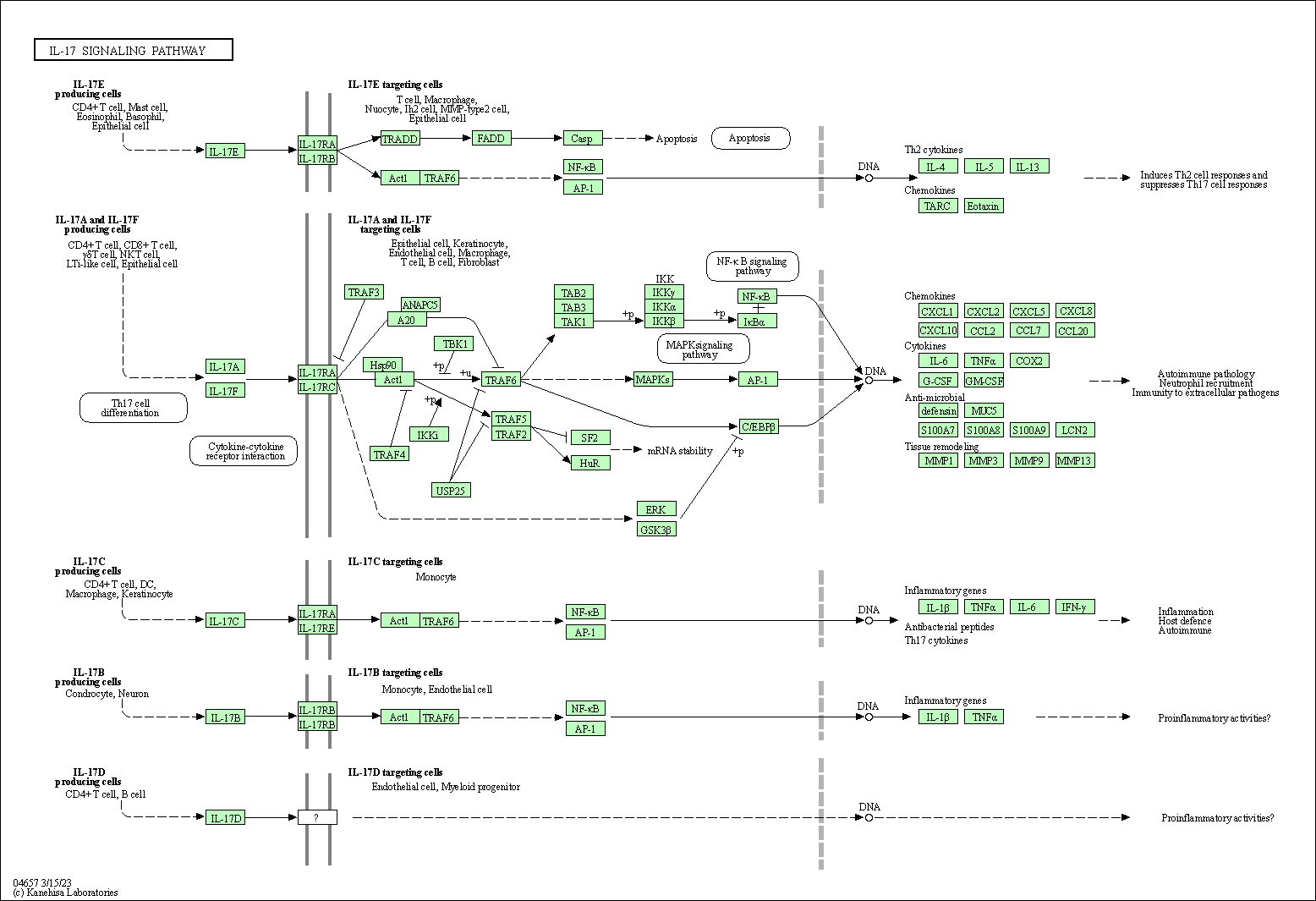
|
|||||||
| Th1 and Th2 cell differentiation | hsa04658 |
Pathway Map 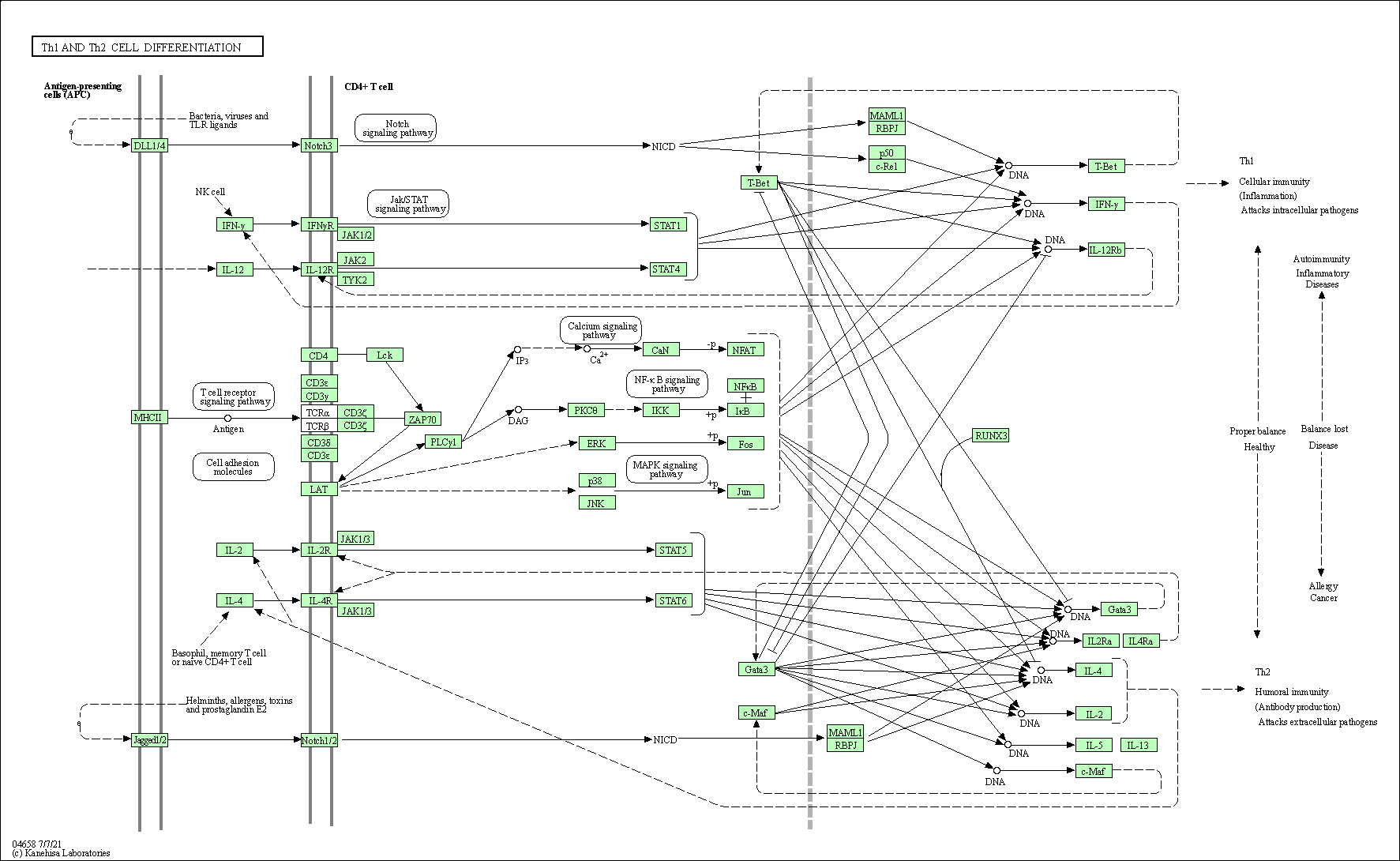
|
|||||||
| Th17 cell differentiation | hsa04659 |
Pathway Map 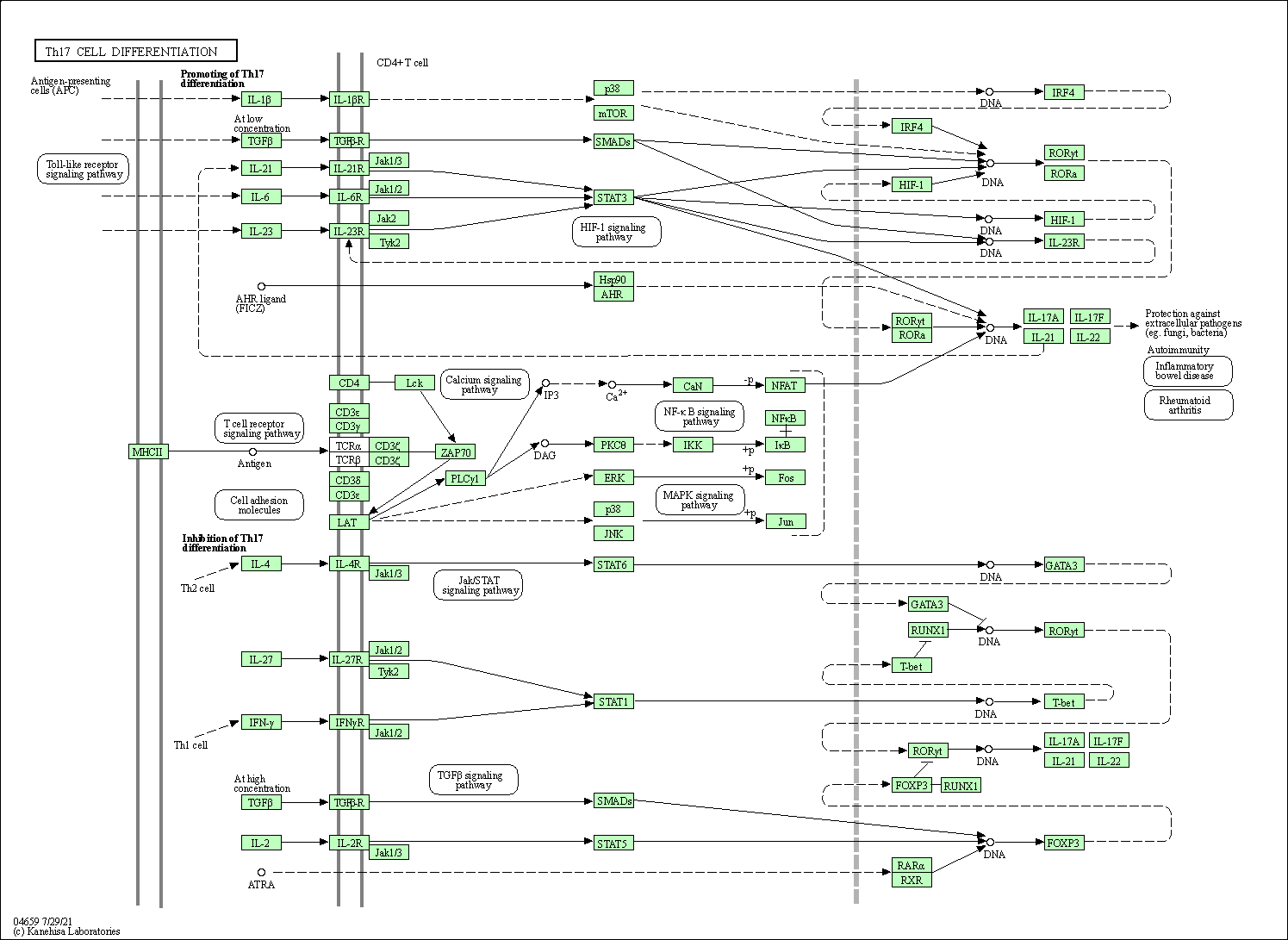
|
|||||||
| T cell receptor signaling pathway | hsa04660 |
Pathway Map 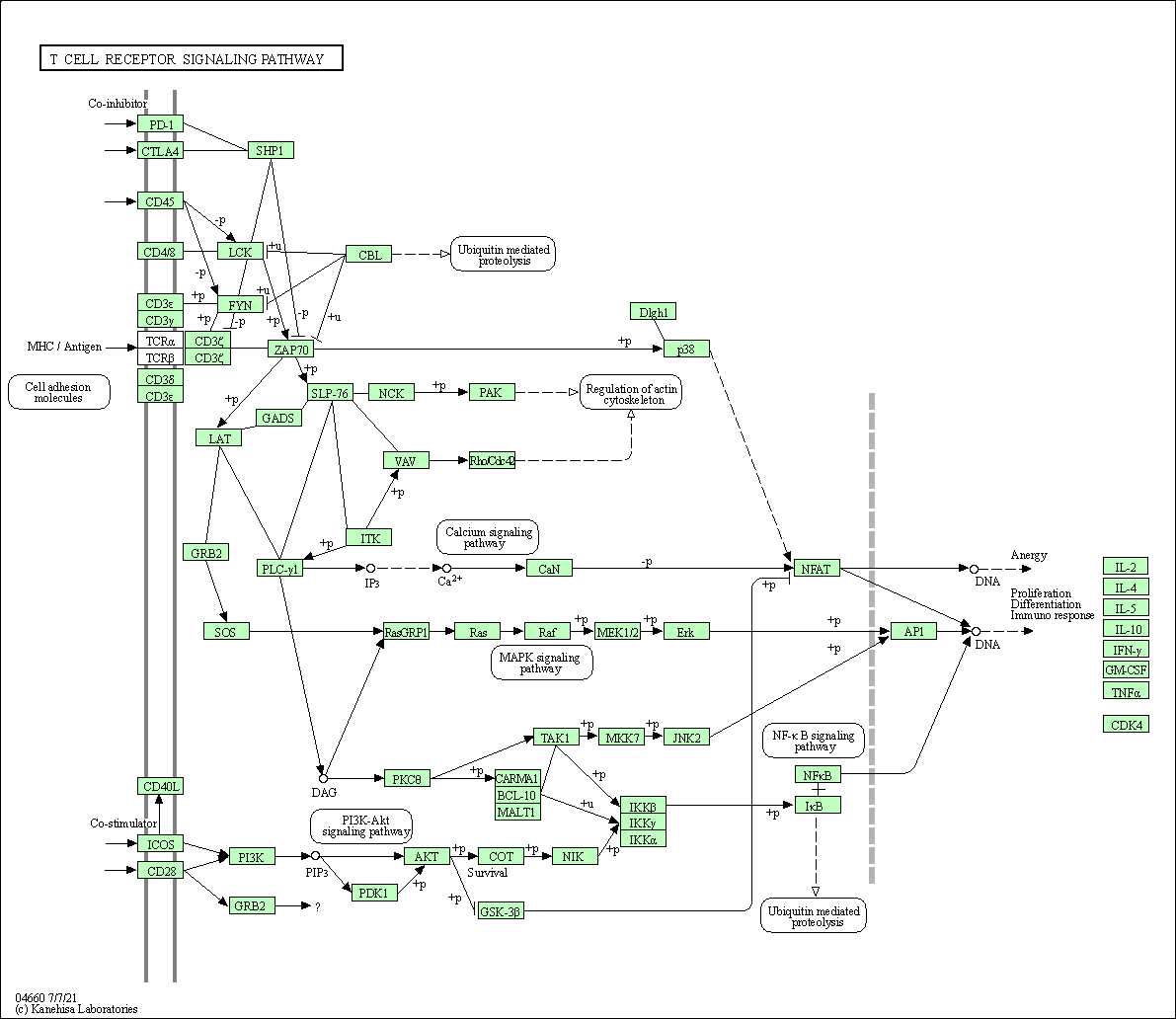
|
|||||||
| B cell receptor signaling pathway | hsa04662 |
Pathway Map 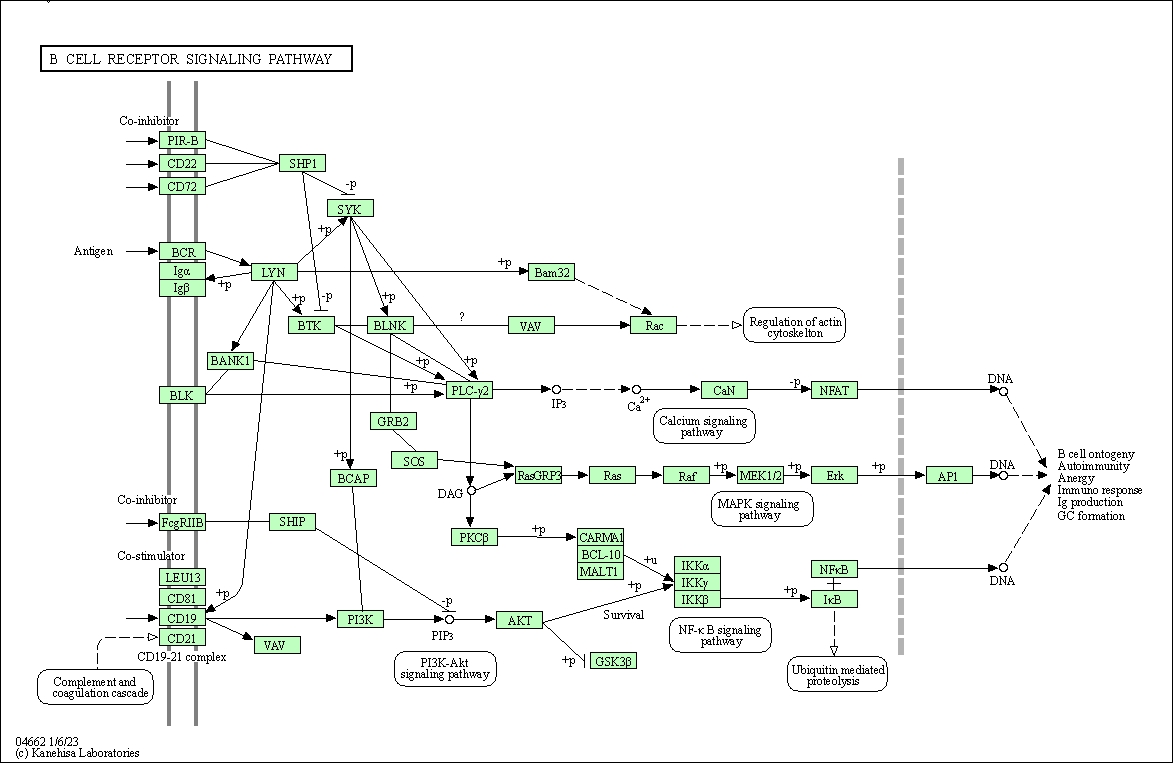
|
|||||||
| Epithelial cell signaling in Helicobacter pylori infection | hsa05120 |
Pathway Map 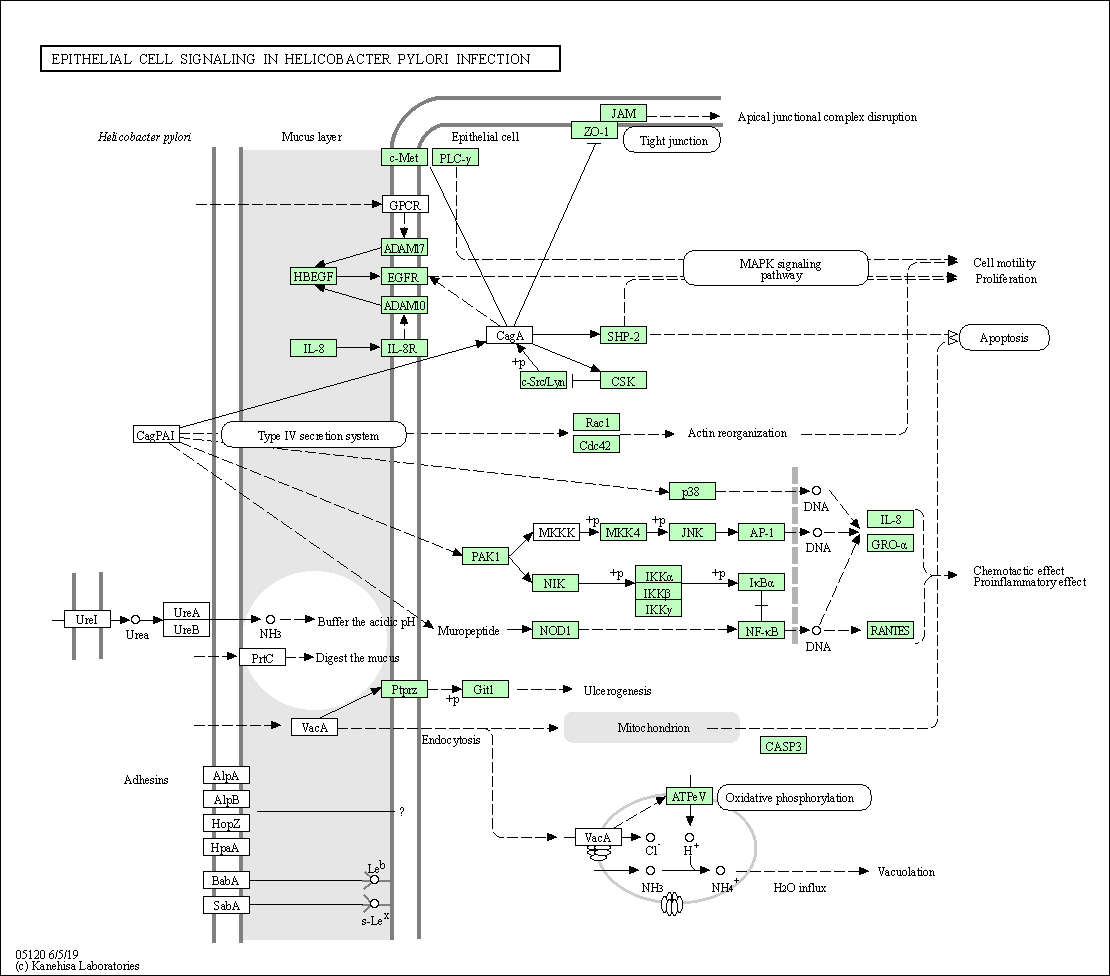
|
|||||||
| Pathogenic Escherichia coli infection | hsa05130 |
Pathway Map 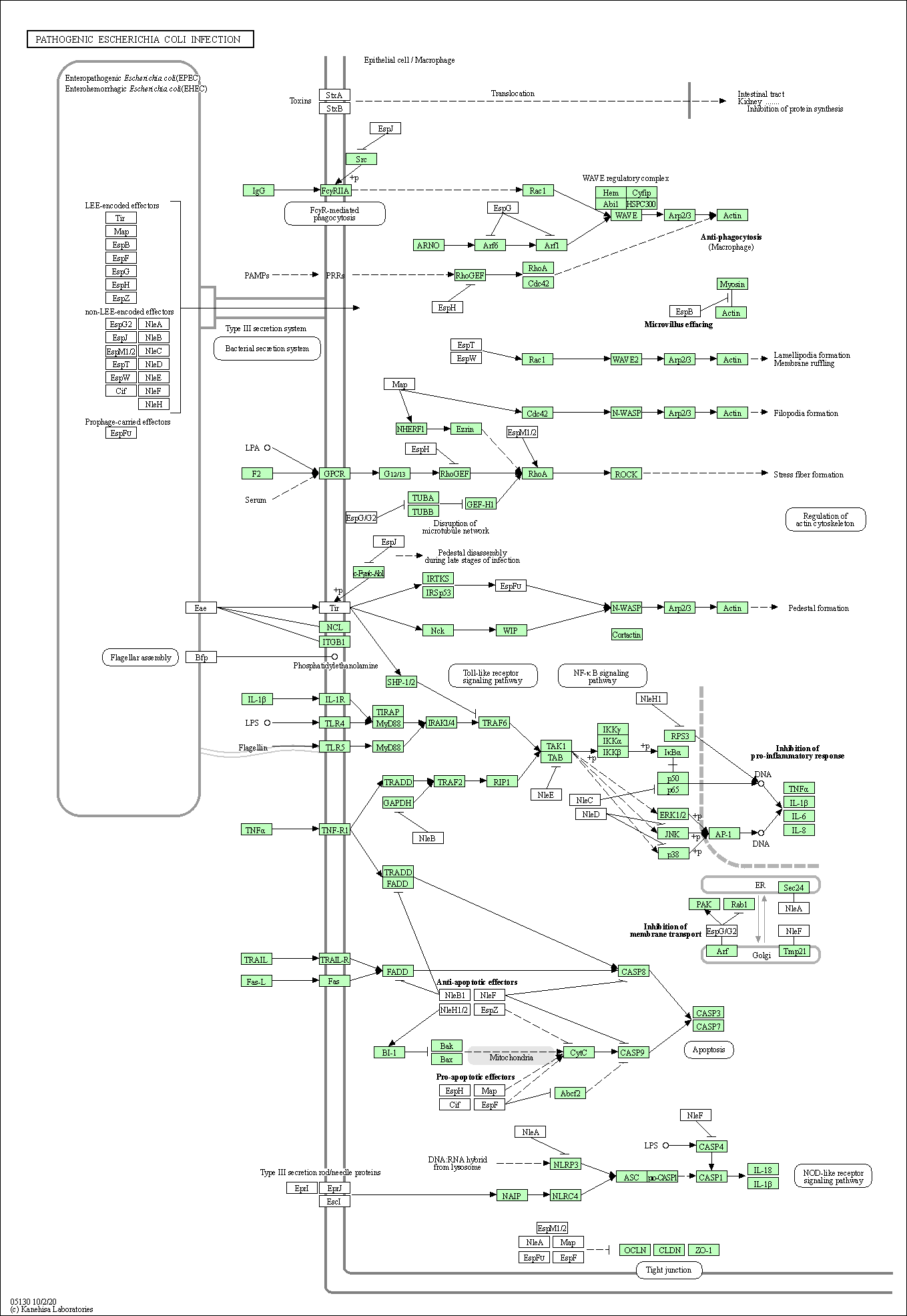
|
|||||||
| Shigellosis | hsa05131 |
Pathway Map 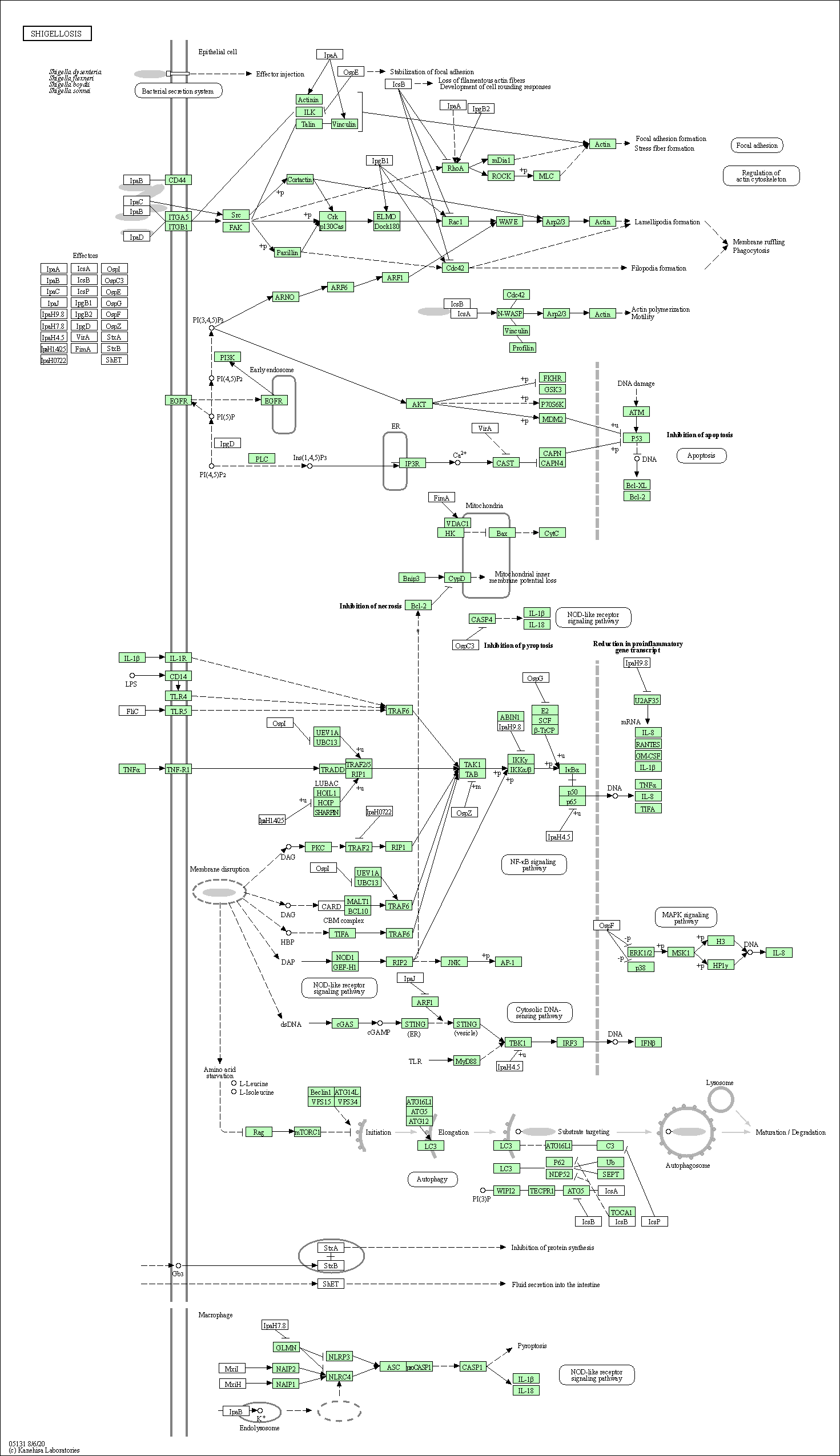
|
|||||||
| Salmonella infection | hsa05132 |
Pathway Map 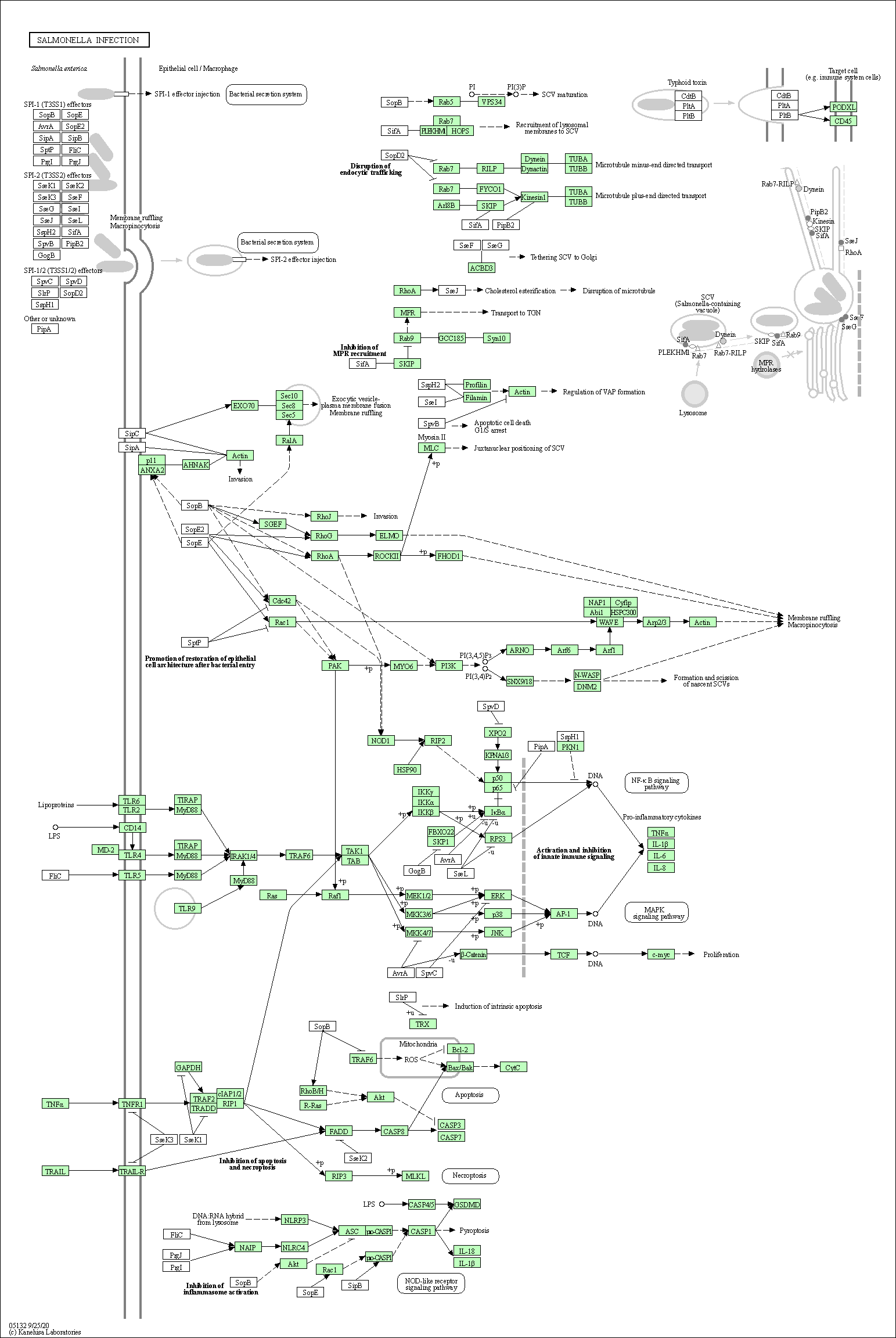
|
|||||||
| Pertussis | hsa05133 |
Pathway Map 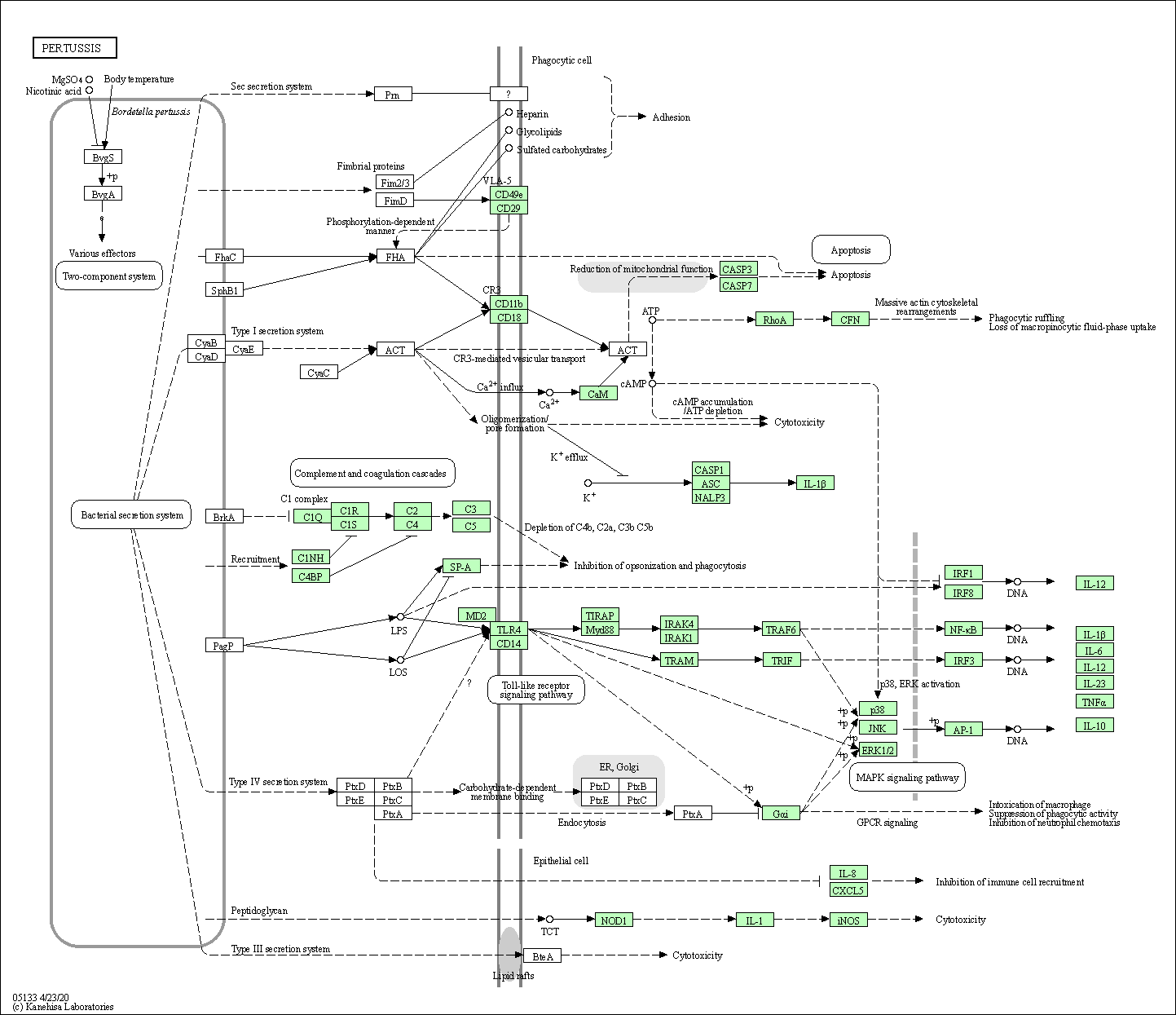
|
|||||||
| Legionellosis | hsa05134 |
Pathway Map 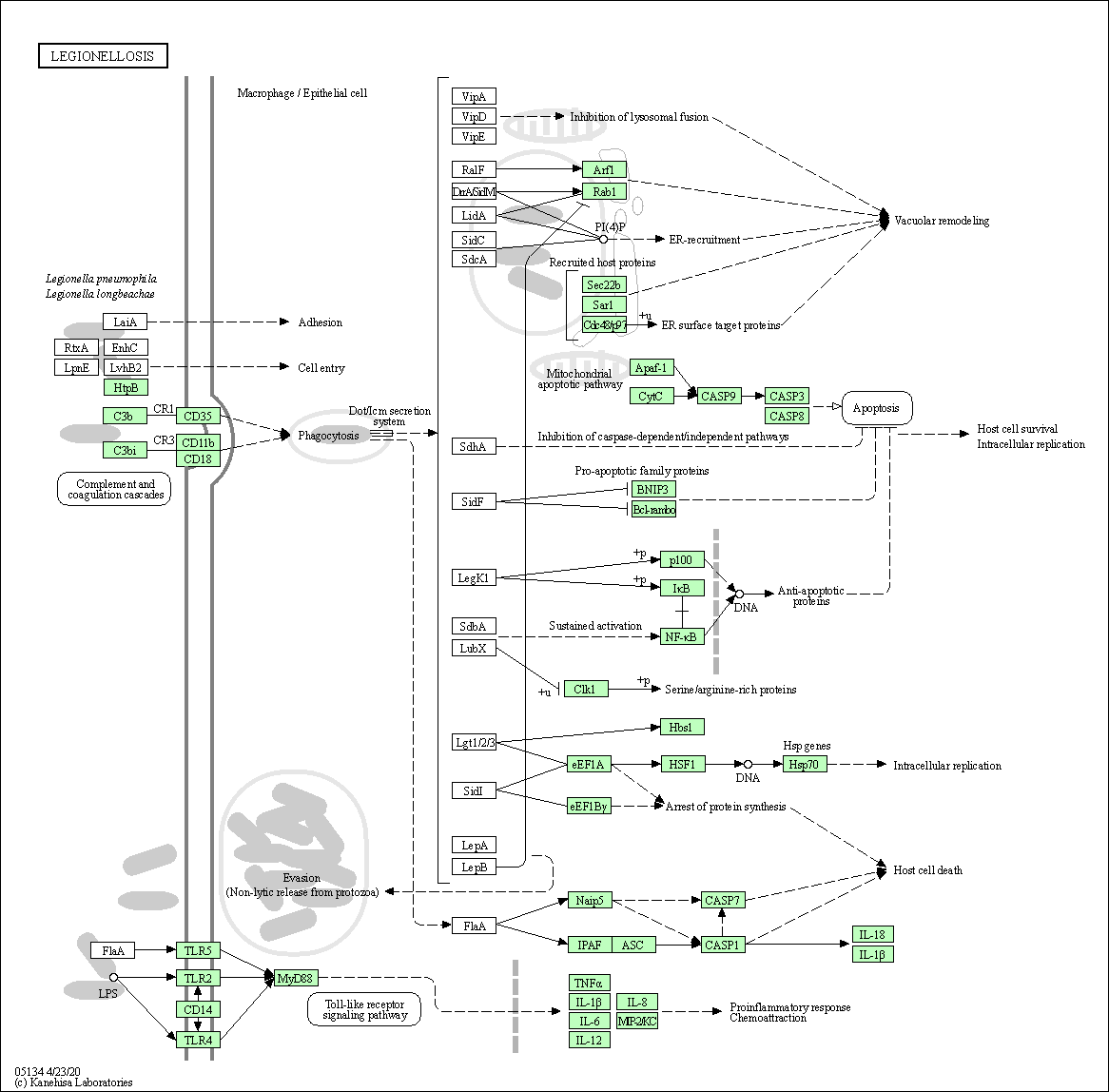
|
|||||||
| Yersinia infection | hsa05135 |
Pathway Map 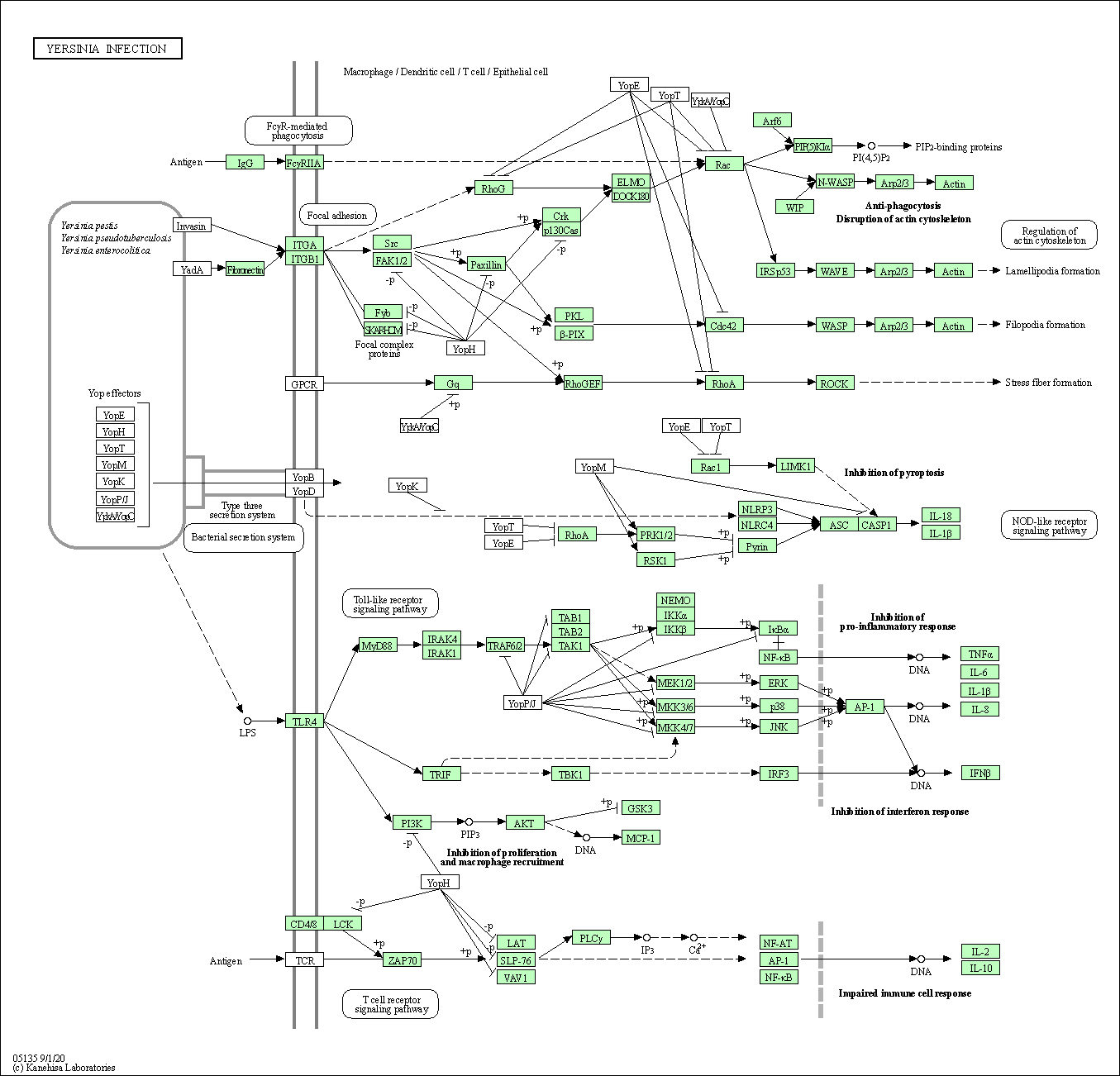
|
|||||||
| Tuberculosis | hsa05152 |
Pathway Map 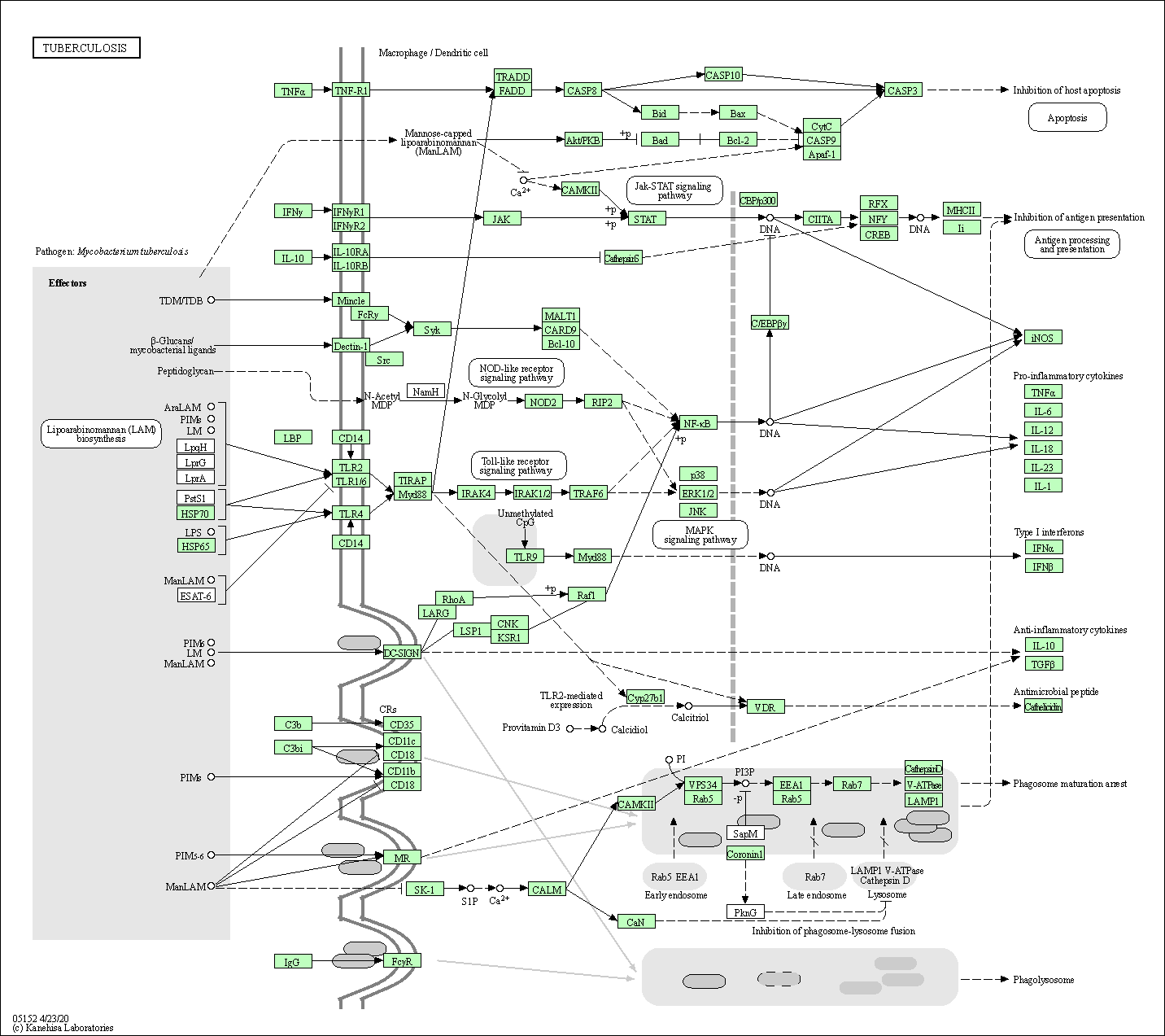
|
|||||||
| Leishmaniasis | hsa05140 |
Pathway Map 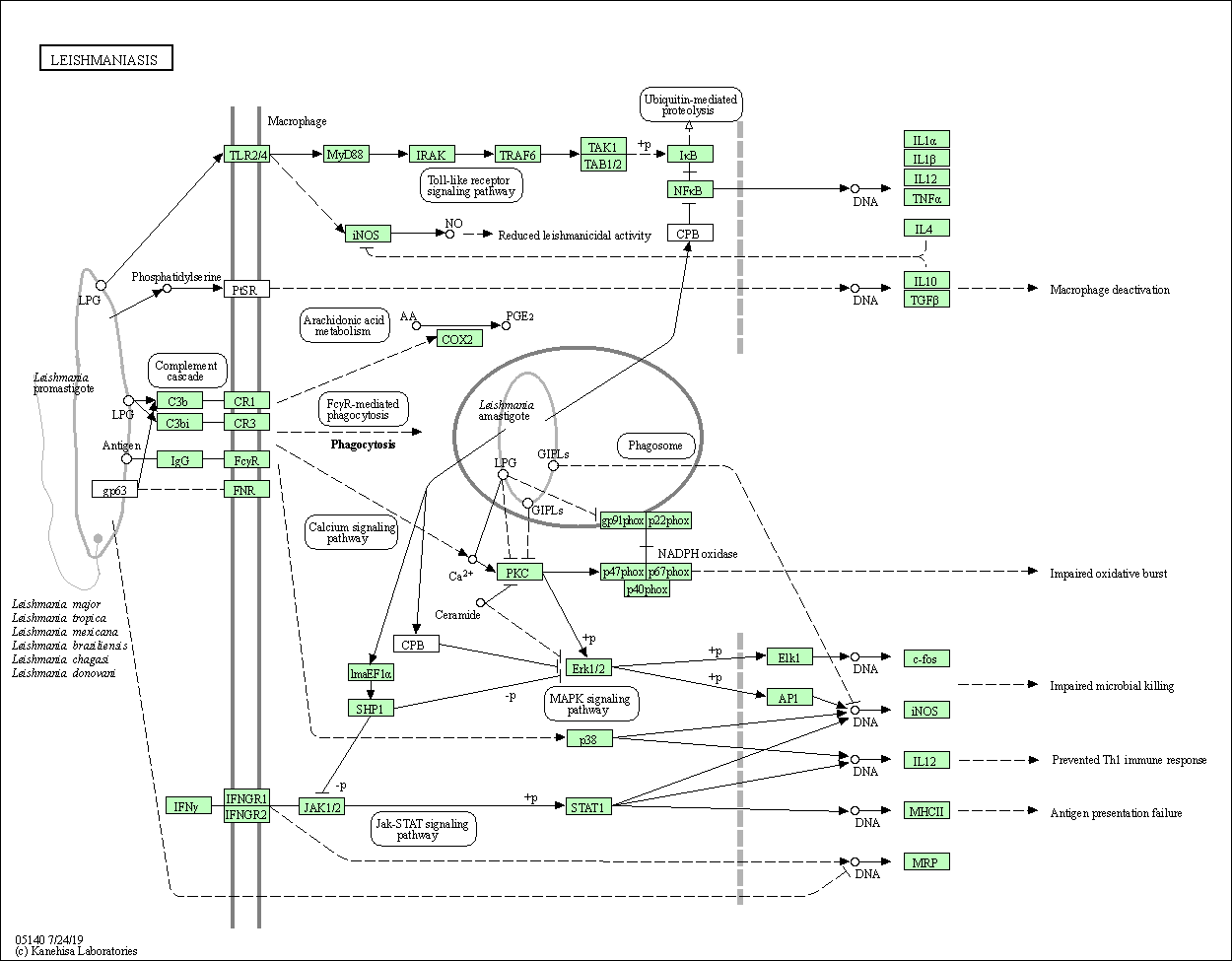
|
|||||||
| Chagas disease | hsa05142 |
Pathway Map 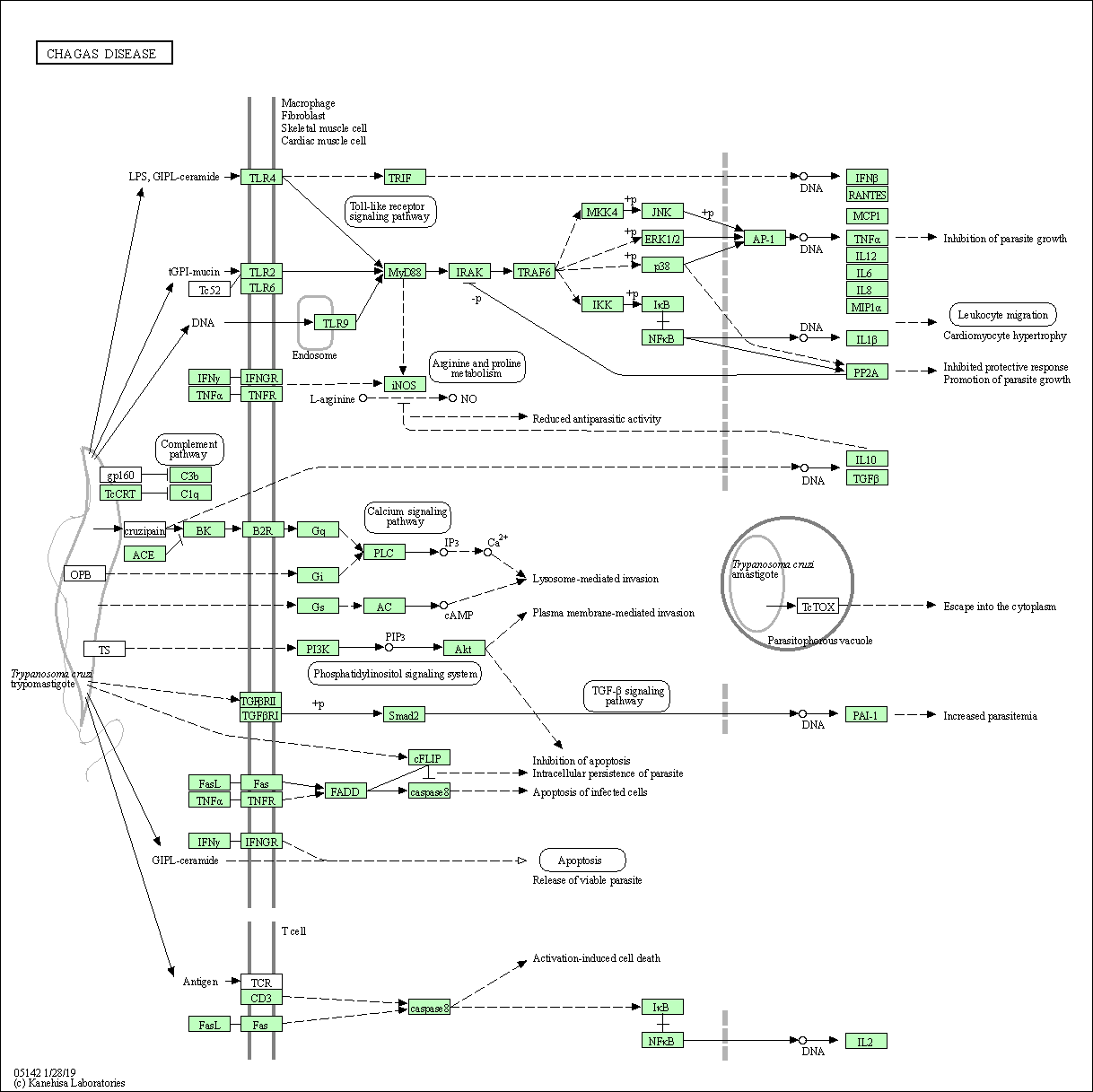
|
|||||||
| Toxoplasmosis | hsa05145 |
Pathway Map 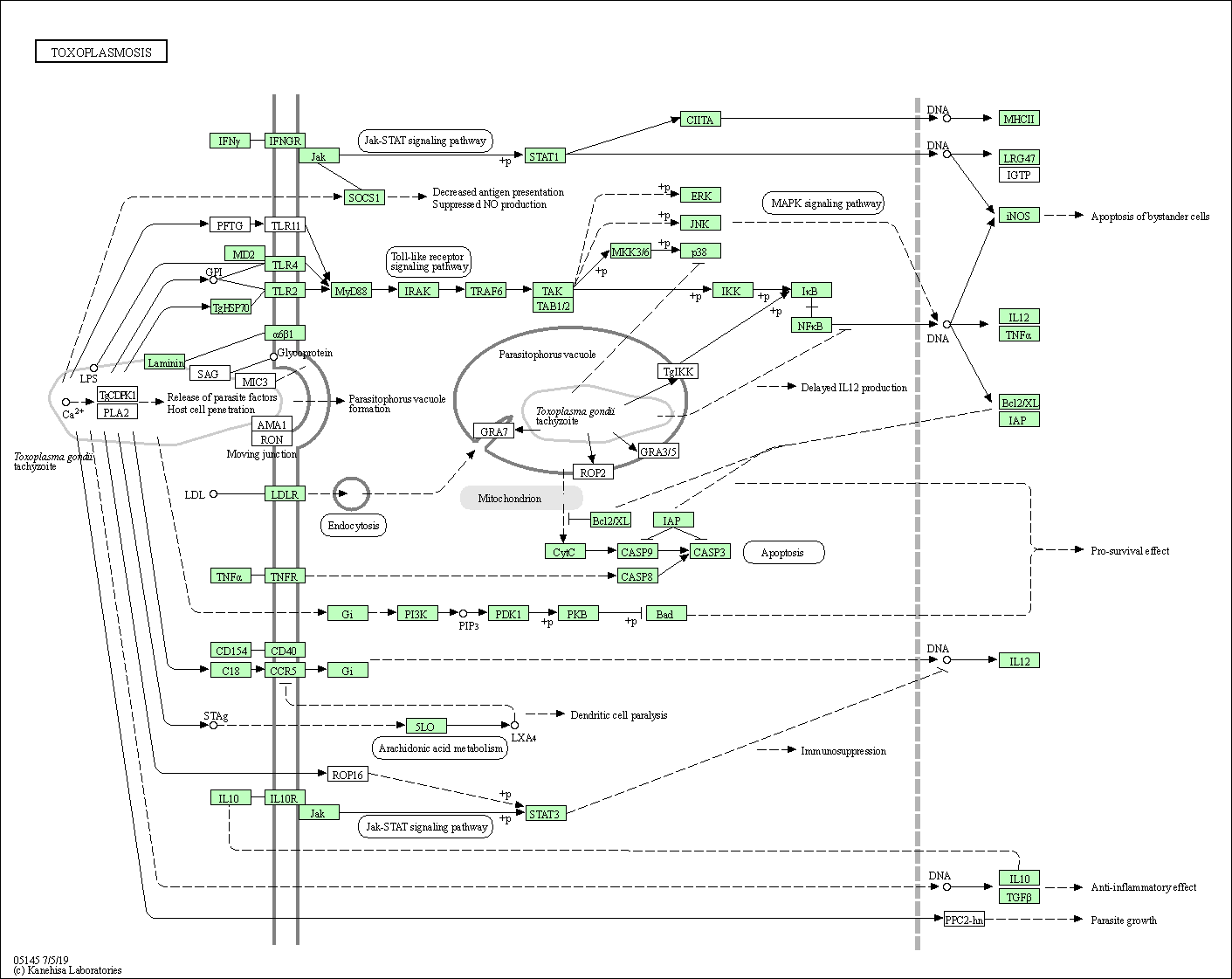
|
|||||||
| Amoebiasis | hsa05146 |
Pathway Map 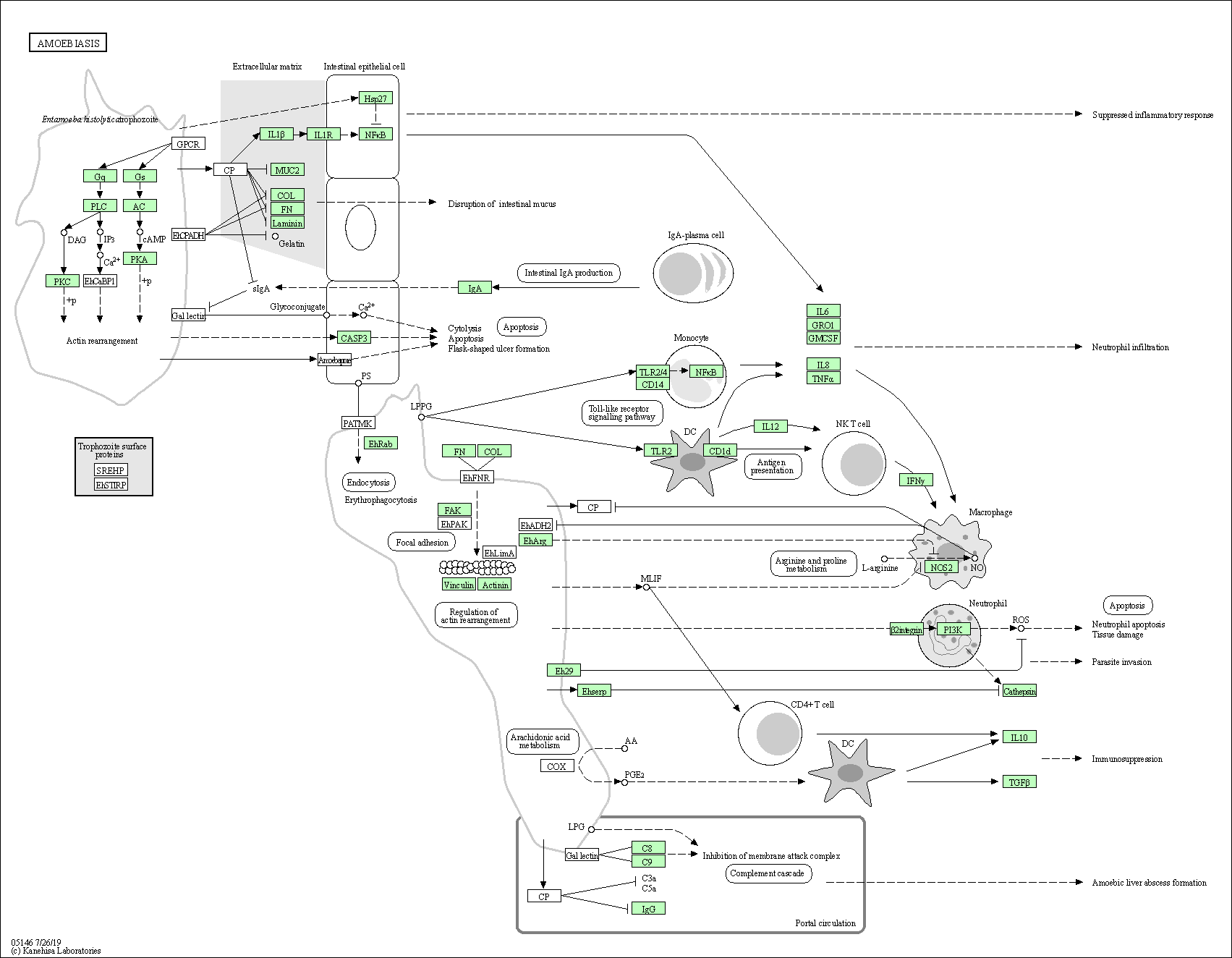
|
|||||||
| Hepatitis C | hsa05160 |
Pathway Map 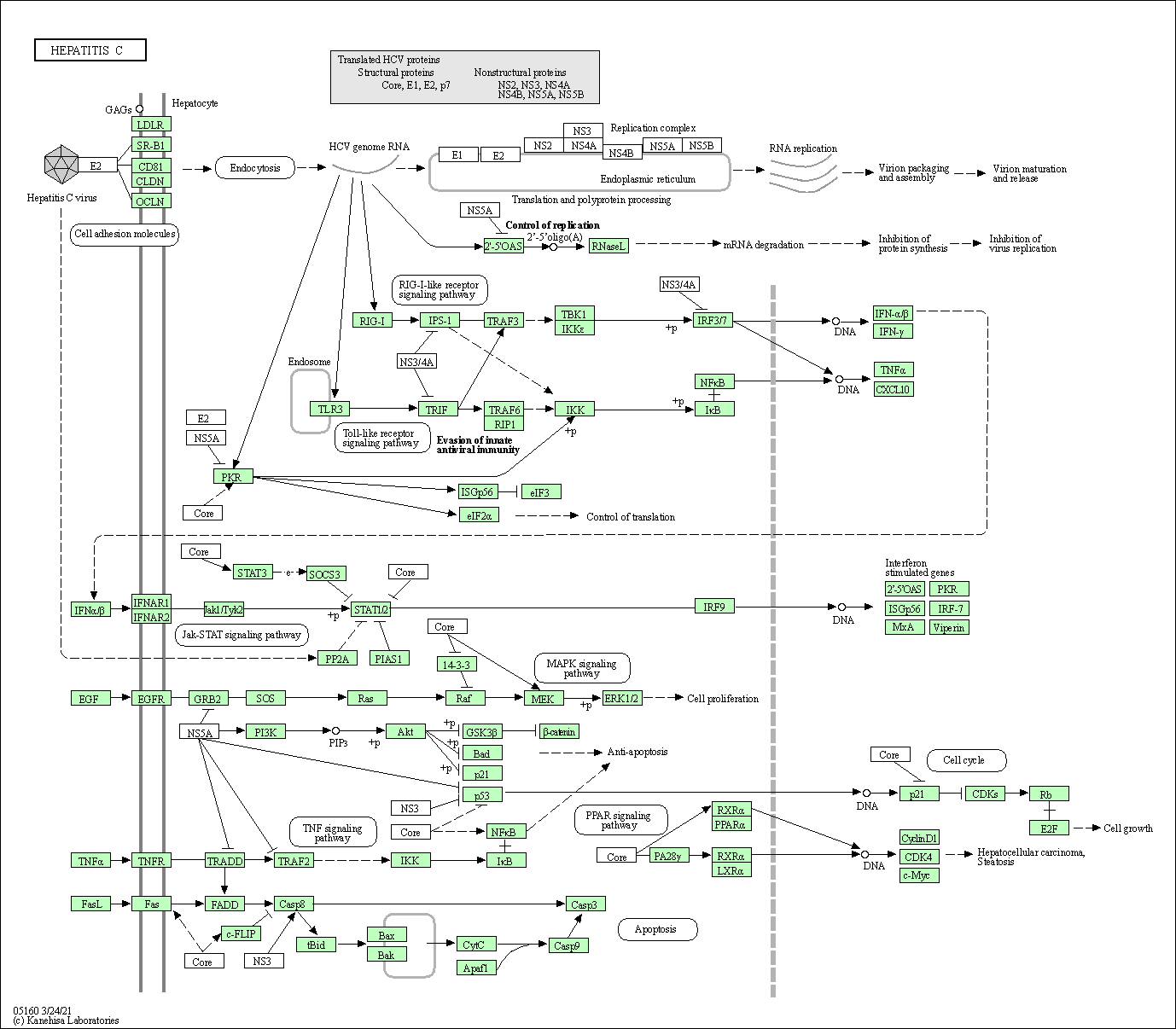
|
|||||||
| Hepatitis B | hsa05161 |
Pathway Map 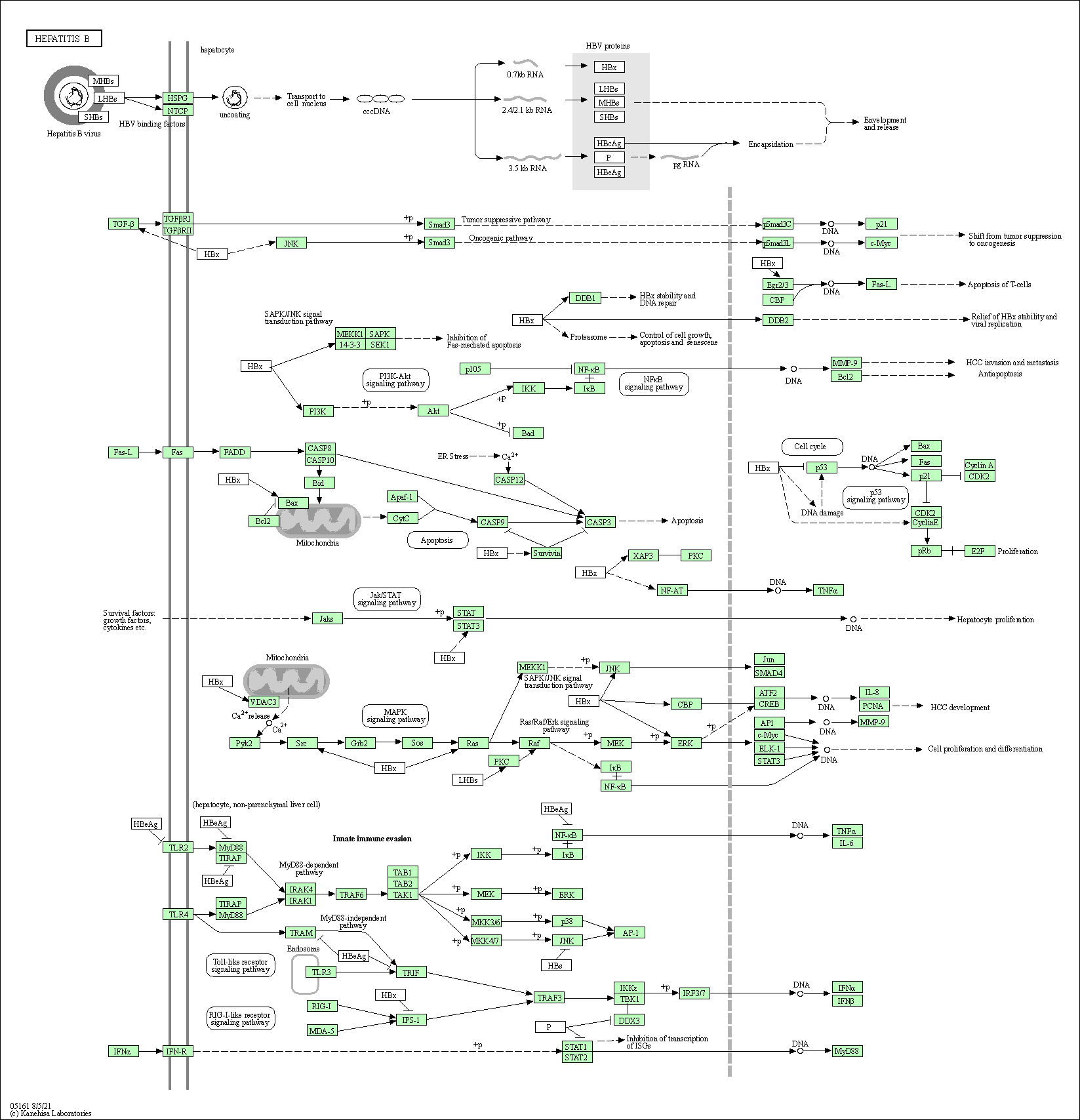
|
|||||||
| Measles | hsa05162 |
Pathway Map 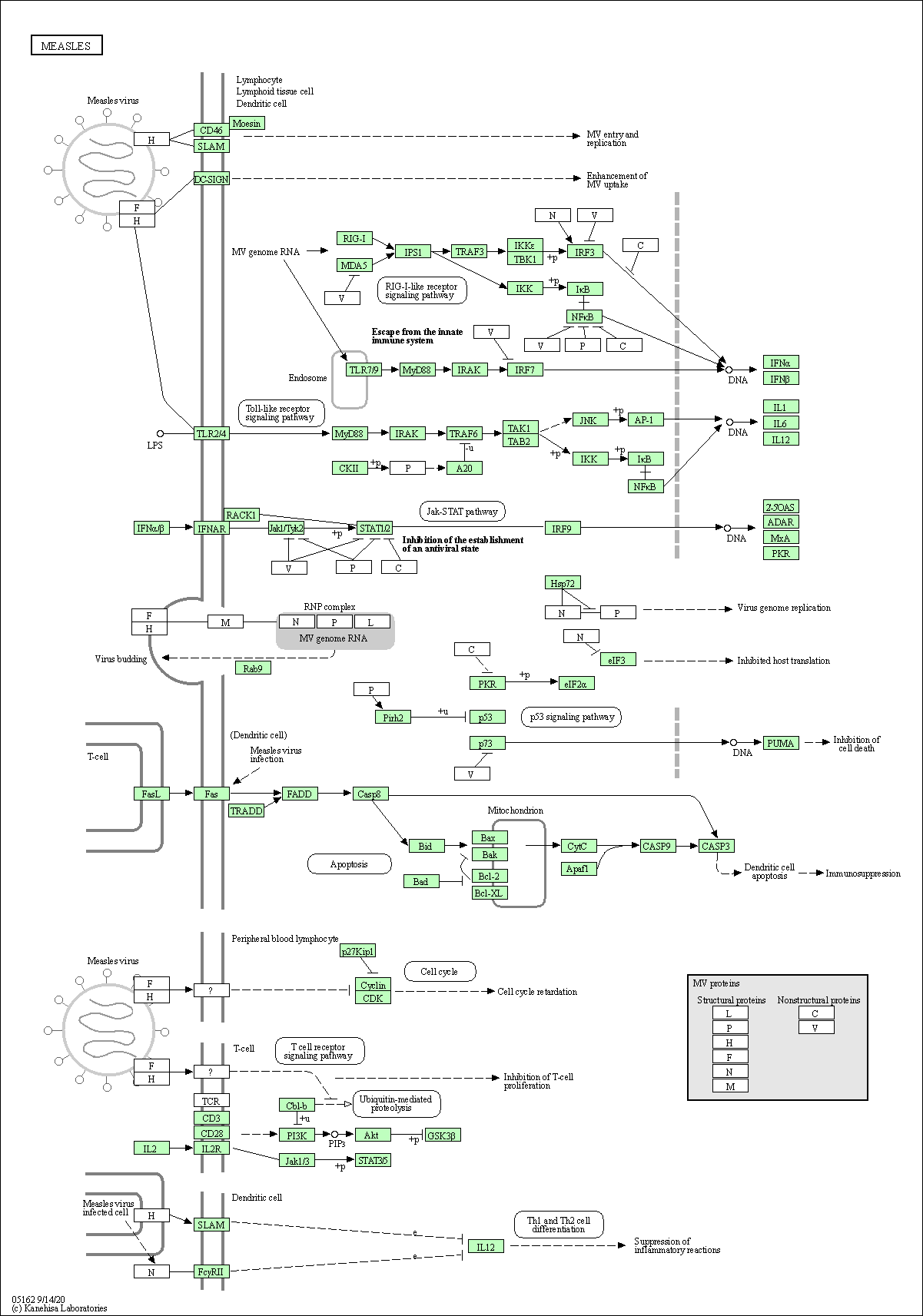
|
|||||||
| Human cytomegalovirus infection | hsa05163 |
Pathway Map 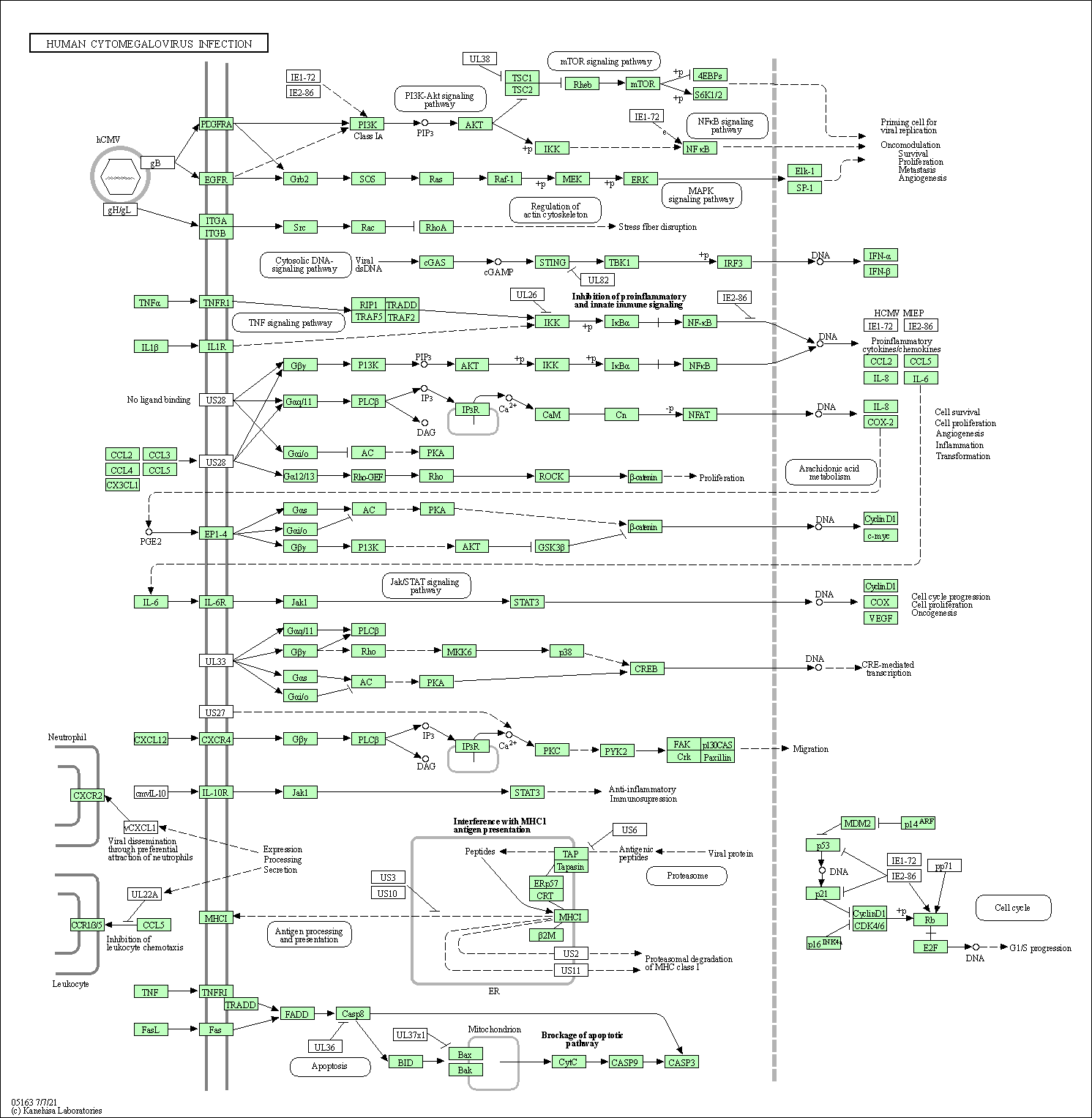
|
|||||||
| Influenza A | hsa05164 |
Pathway Map 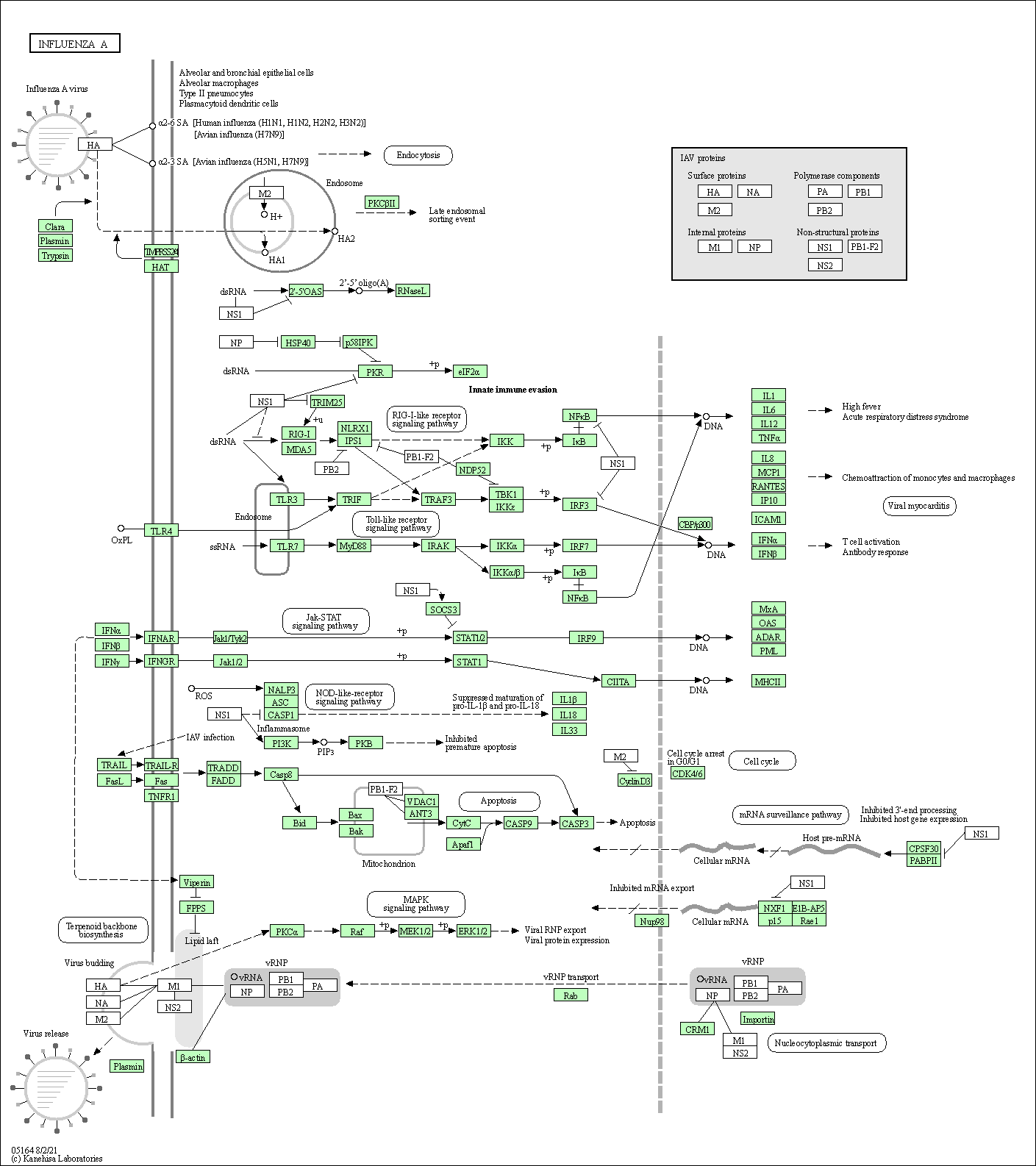
|
|||||||
| Human papillomavirus infection | hsa05165 |
Pathway Map 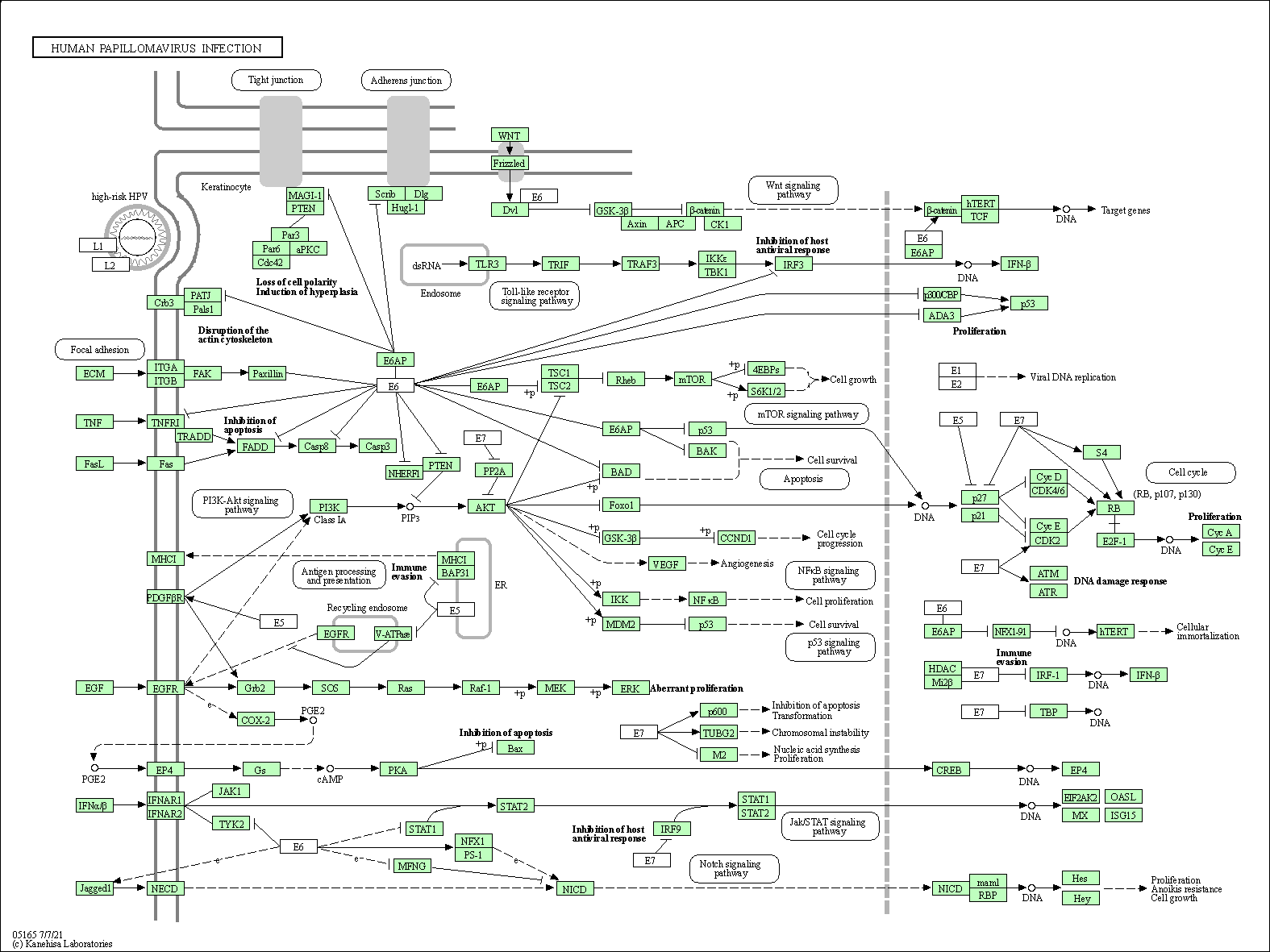
|
|||||||
| Human T-cell leukemia virus 1 infection | hsa05166 |
Pathway Map 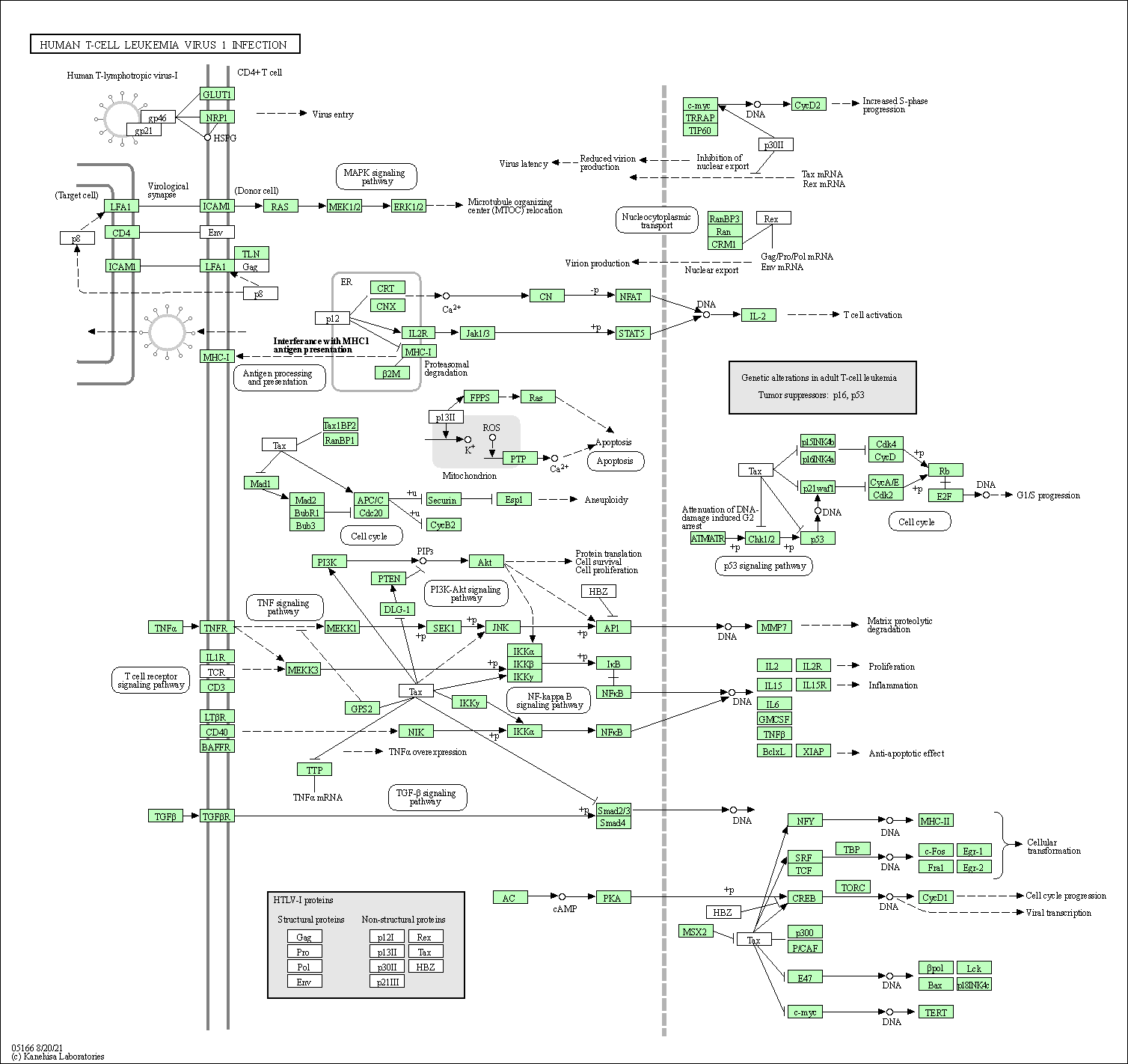
|
|||||||
| Kaposi sarcoma-associated herpesvirus infection | hsa05167 |
Pathway Map 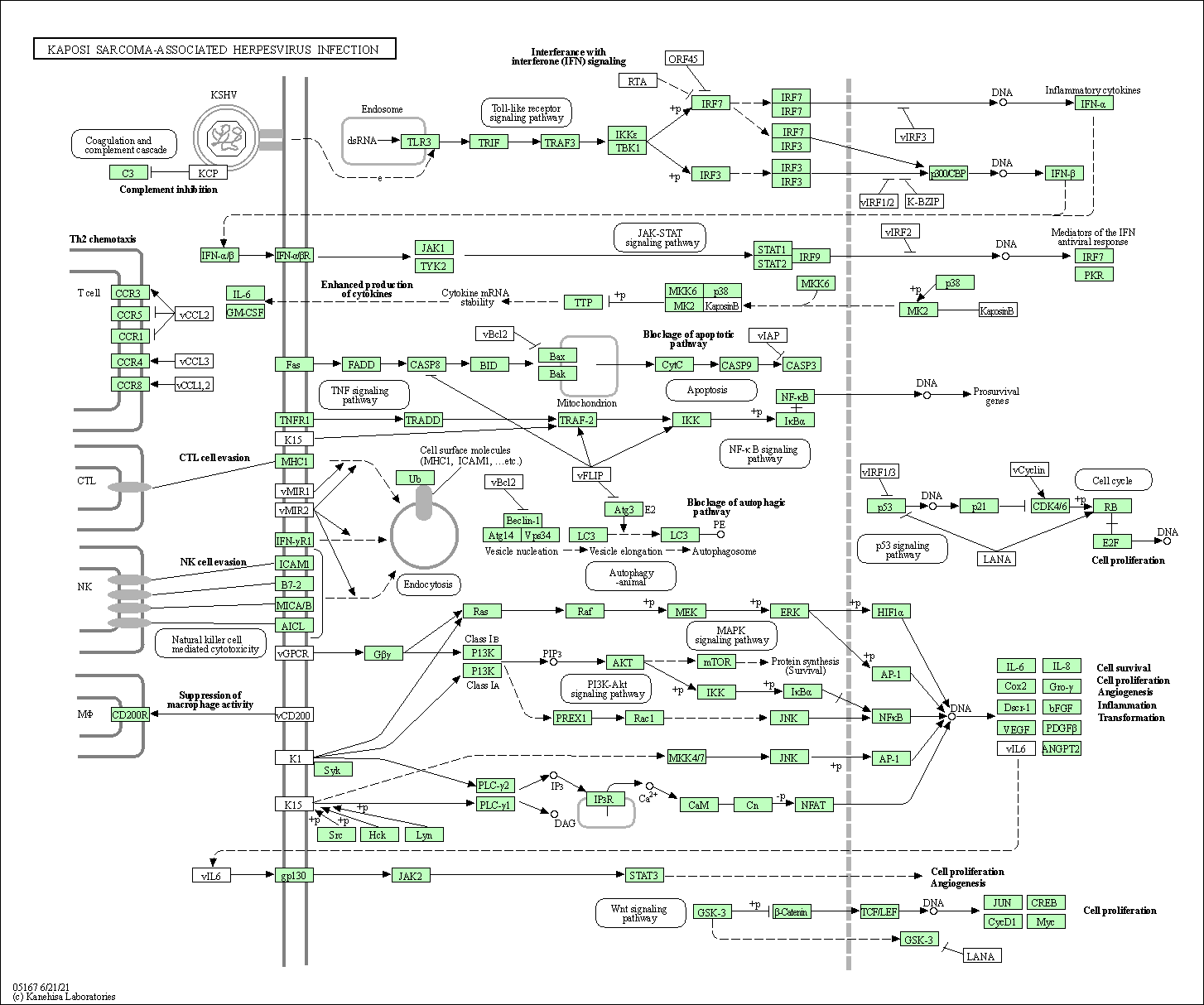
|
|||||||
| Herpes simplex virus 1 infection | hsa05168 |
Pathway Map 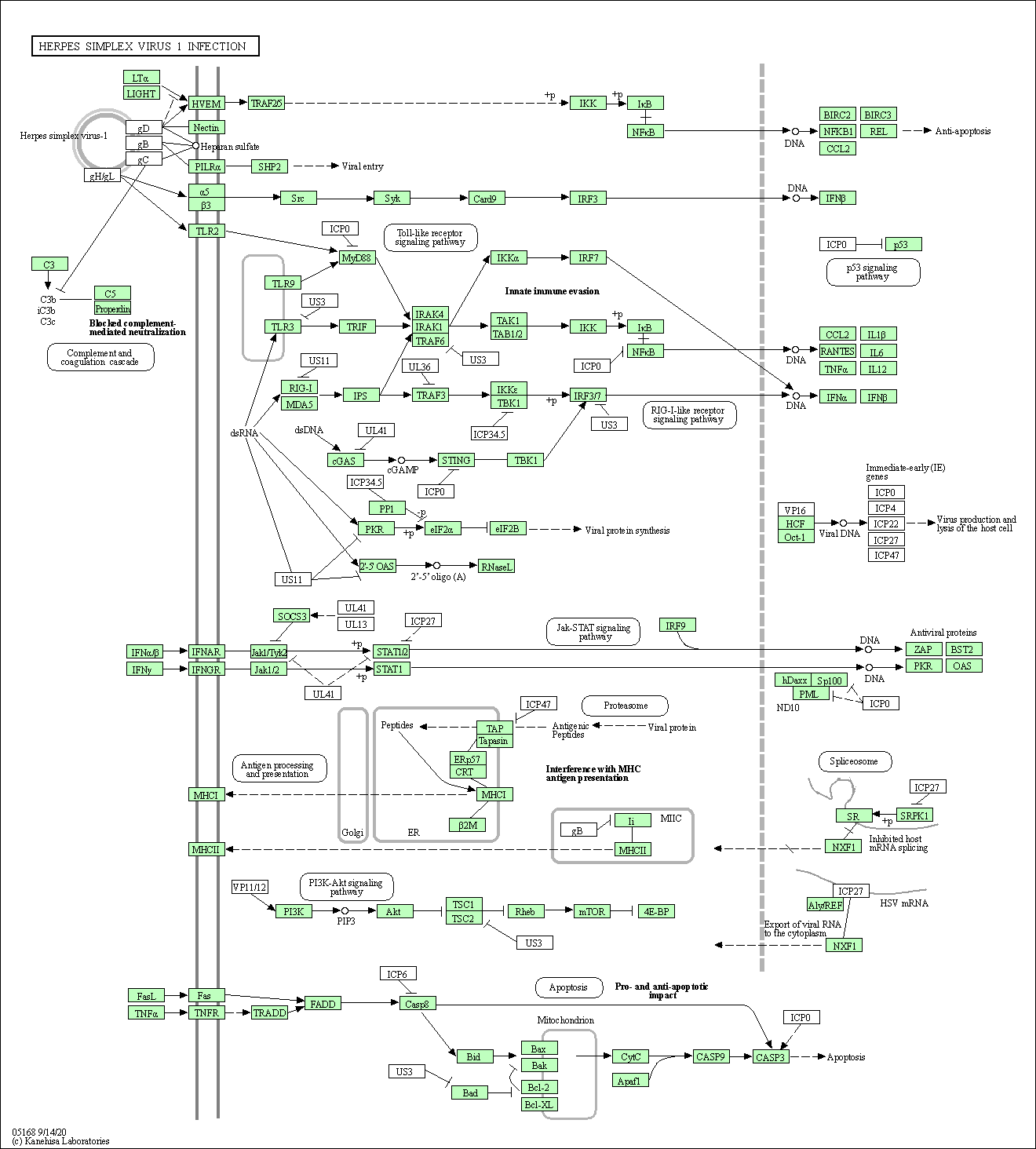
|
|||||||
| Epstein-Barr virus infection | hsa05169 |
Pathway Map 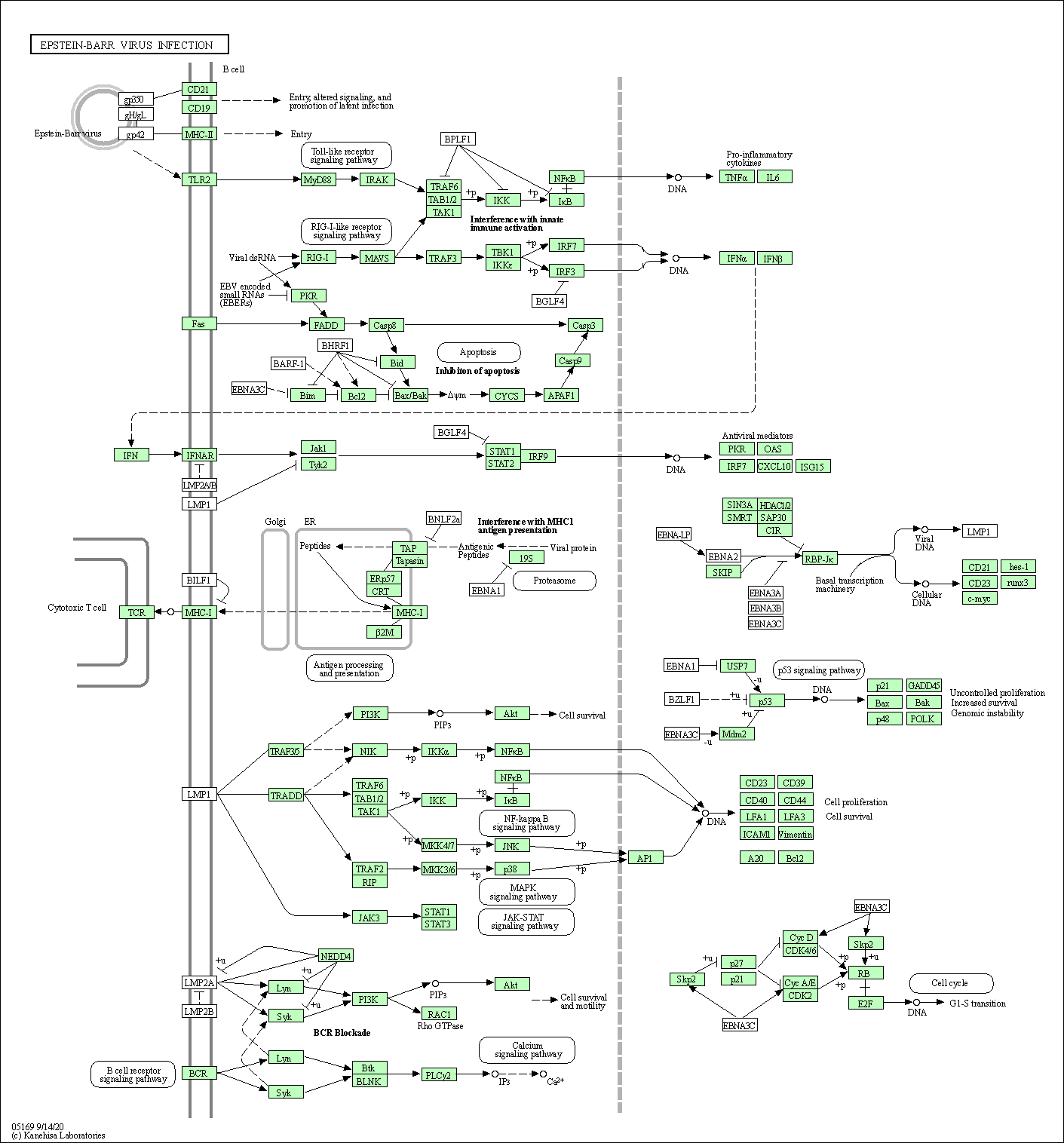
|
|||||||
| Human immunodeficiency virus 1 infection | hsa05170 |
Pathway Map 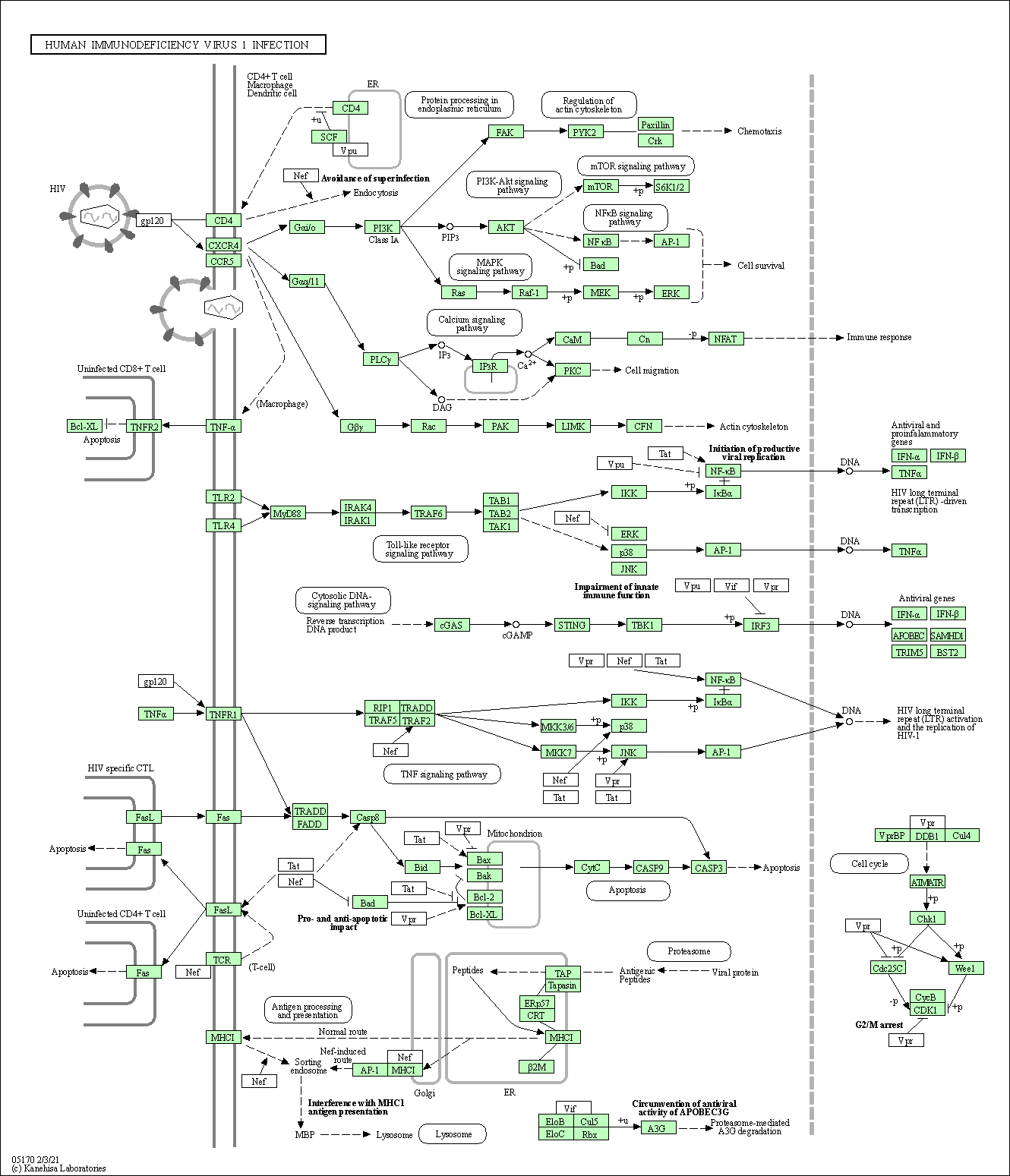
|
|||||||
| Coronavirus disease - COVID-19 | hsa05171 |
Pathway Map 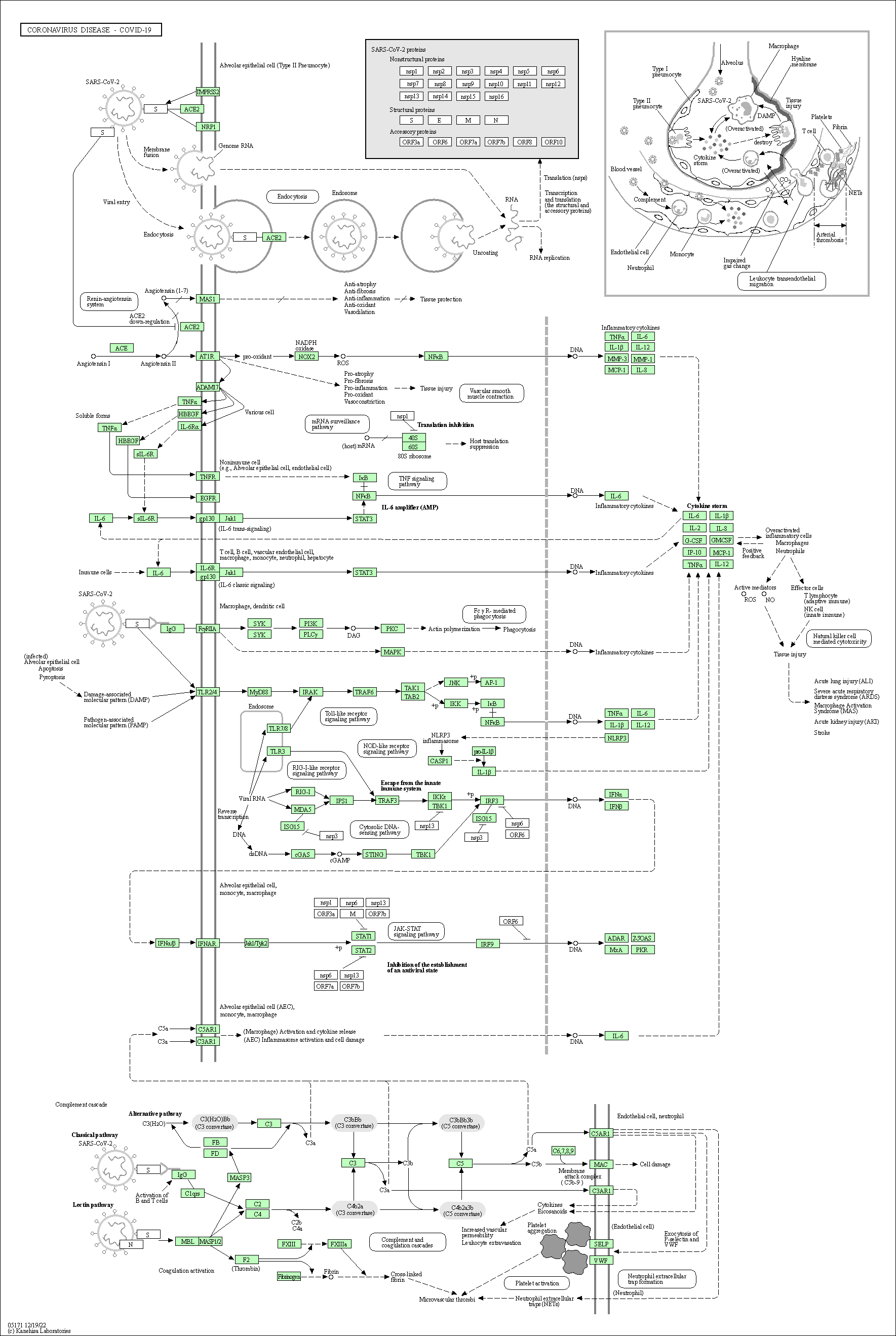
|
|||||||
| Antifolate resistance | hsa01523 |
Pathway Map 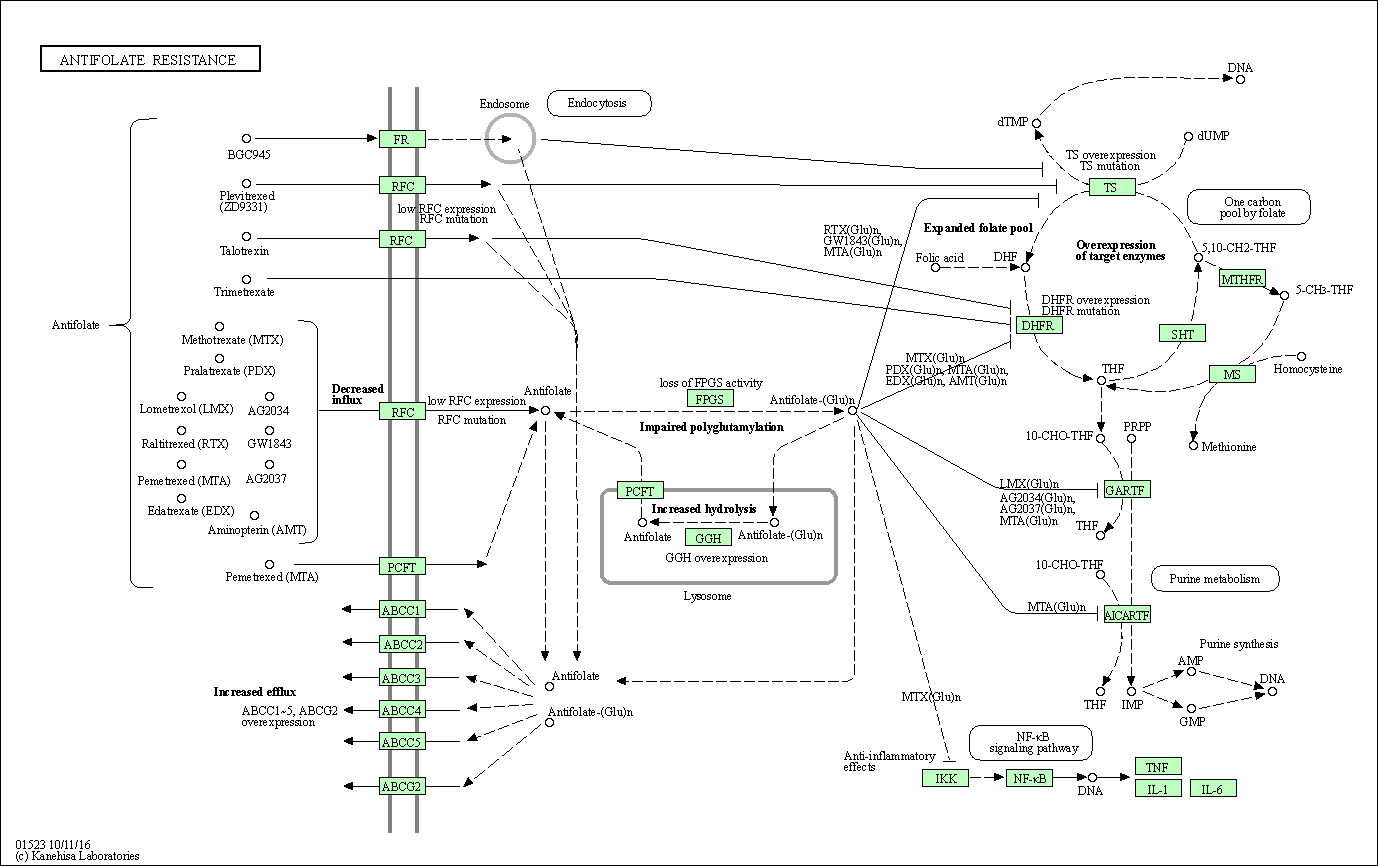
|
|||||||
| Longevity regulating pathway | hsa04211 |
Pathway Map 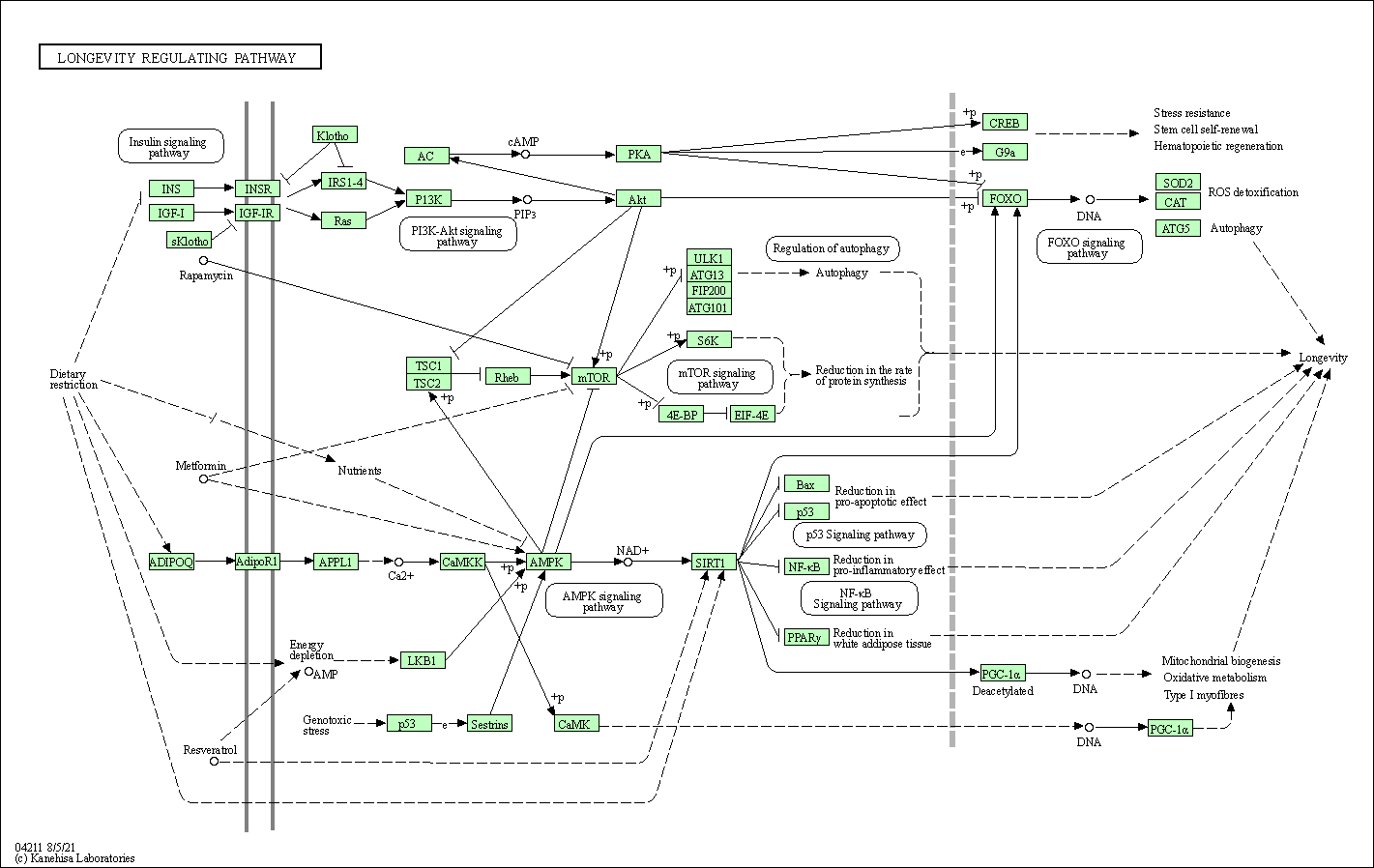
|
|||||||
| Pathways in cancer | hsa05200 |
Pathway Map 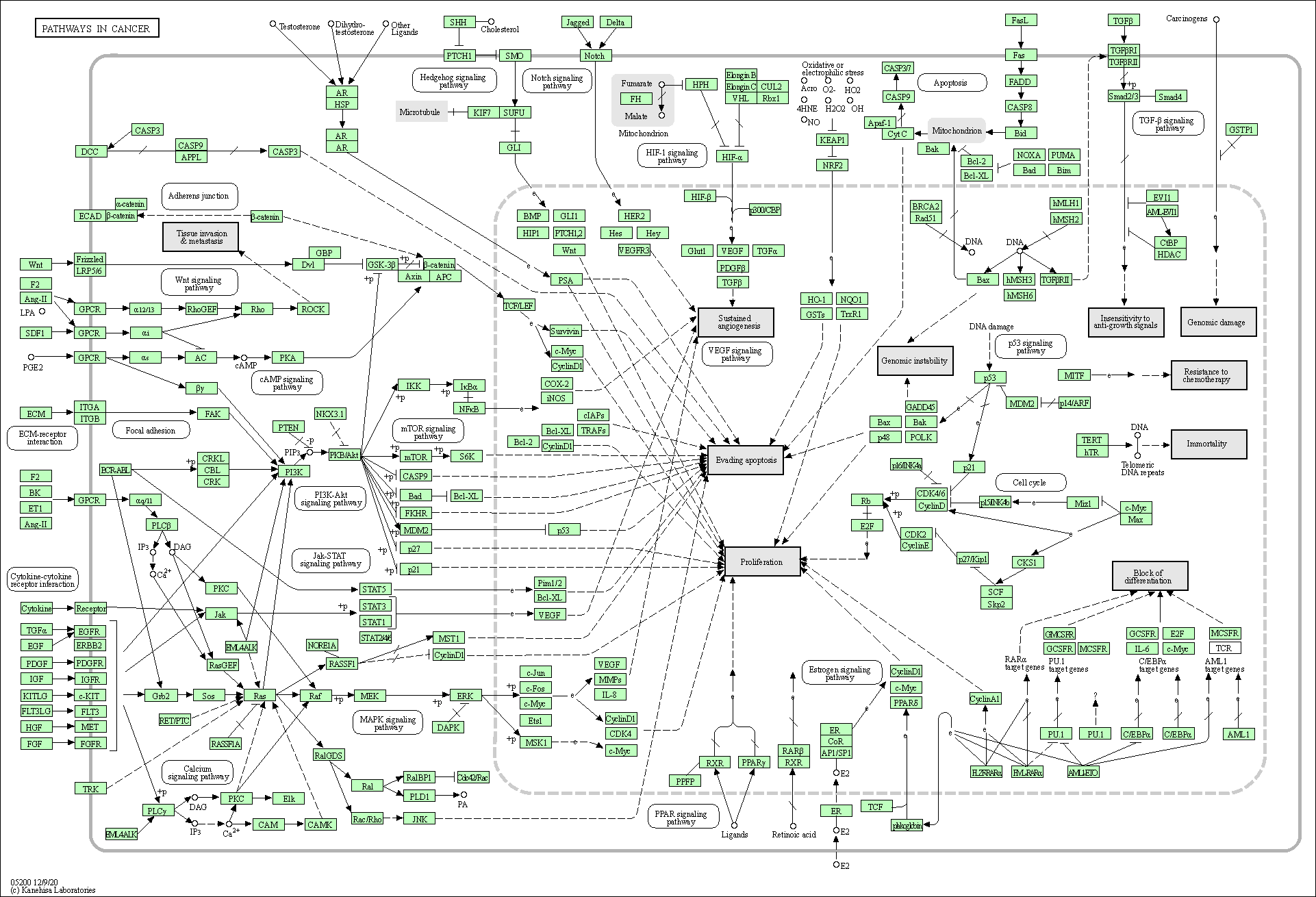
|
|||||||
| Transcriptional misregulation in cancer | hsa05202 |
Pathway Map 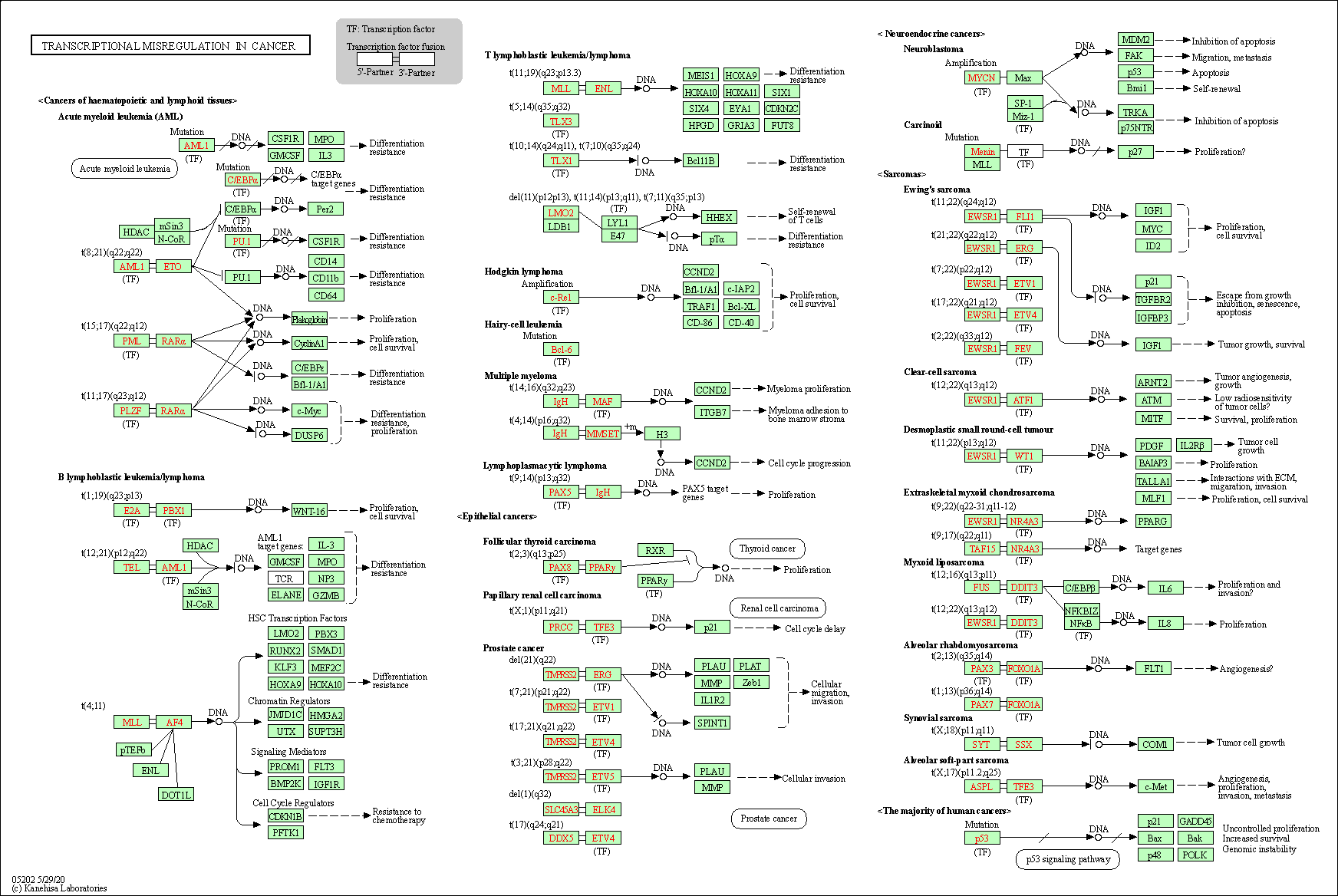
|
|||||||
| Viral carcinogenesis | hsa05203 |
Pathway Map 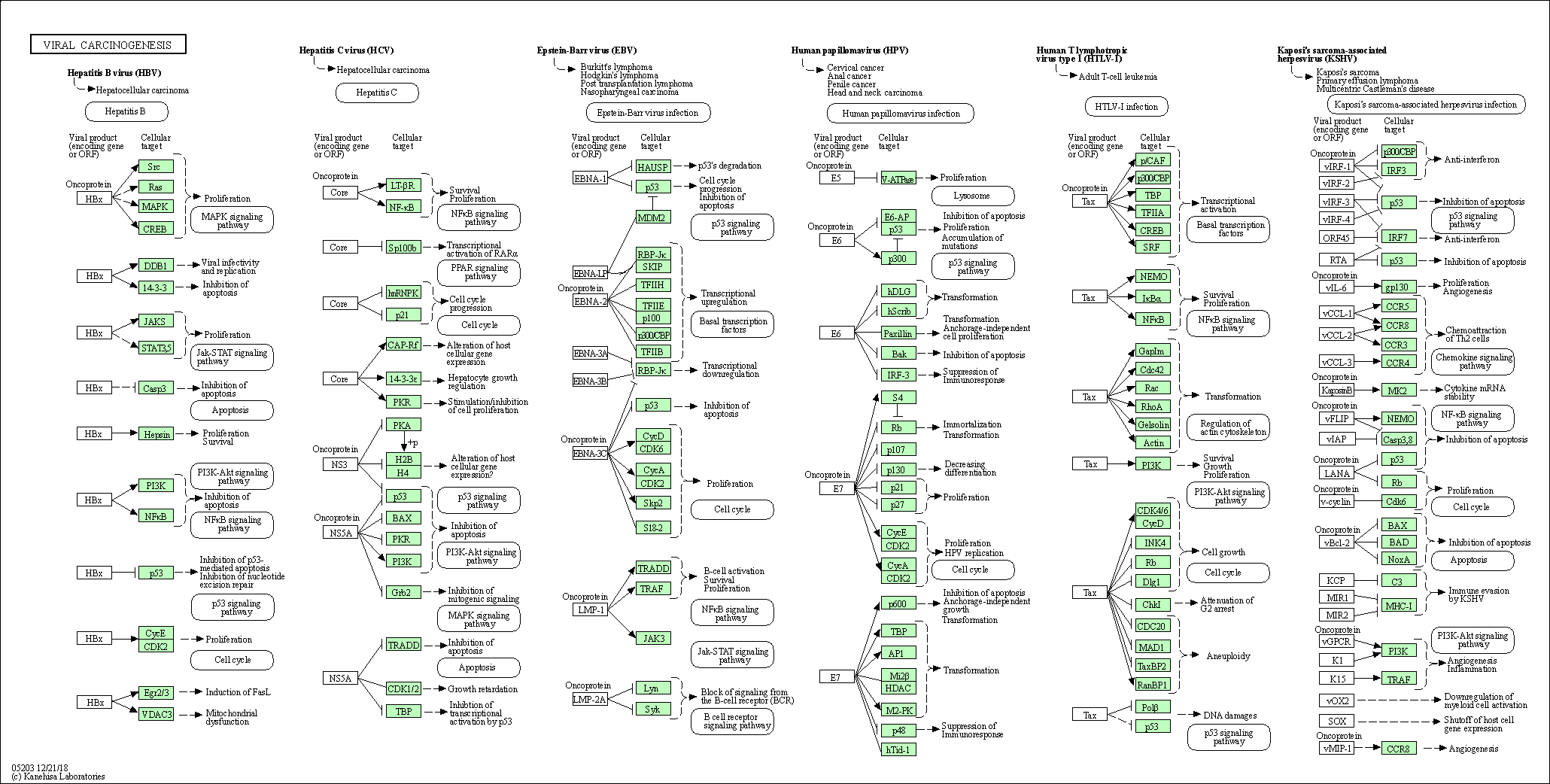
|
|||||||
| Chemical carcinogenesis - receptor activation | hsa05207 |
Pathway Map 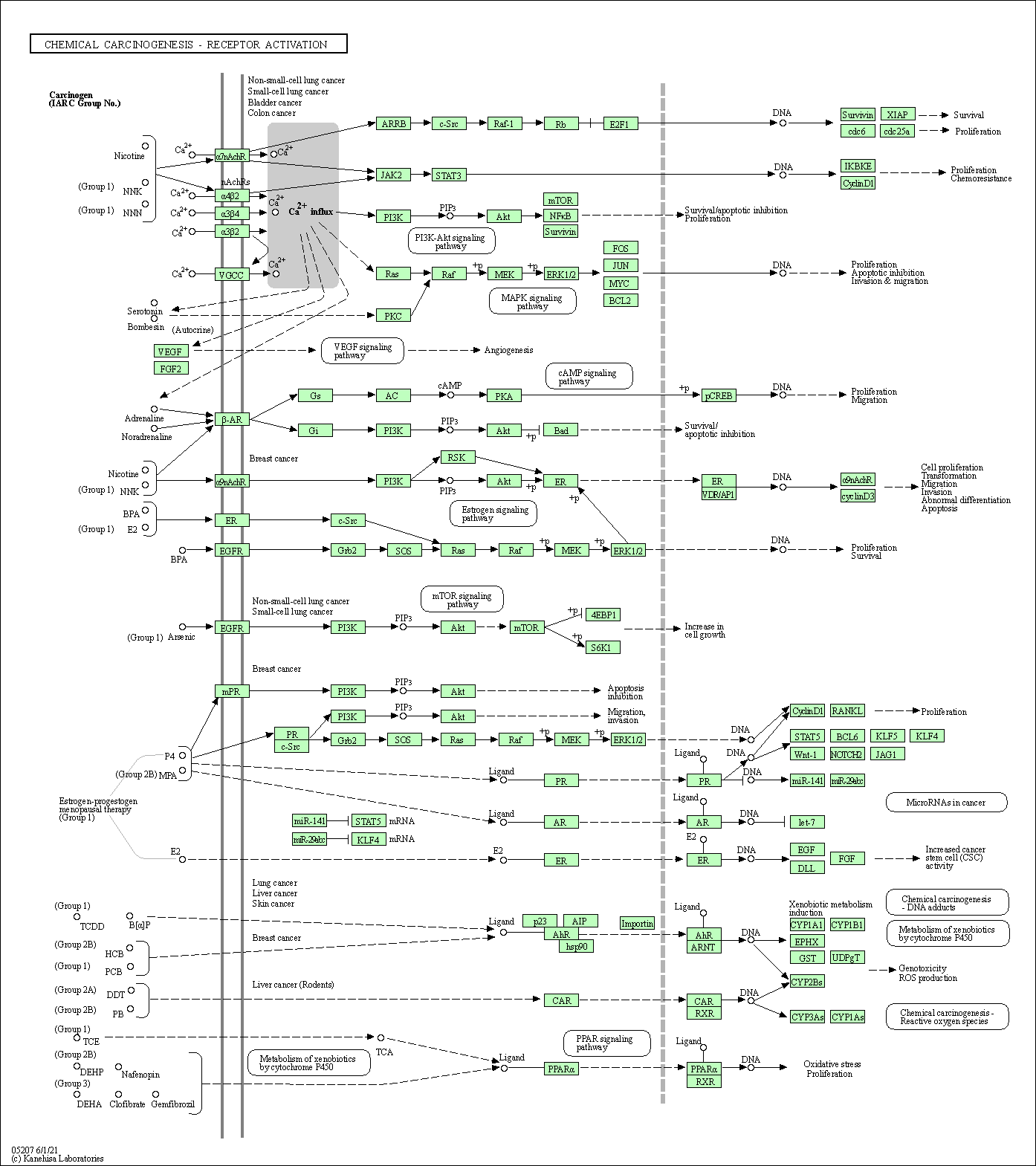
|
|||||||
| Chemical carcinogenesis - reactive oxygen species | hsa05208 |
Pathway Map 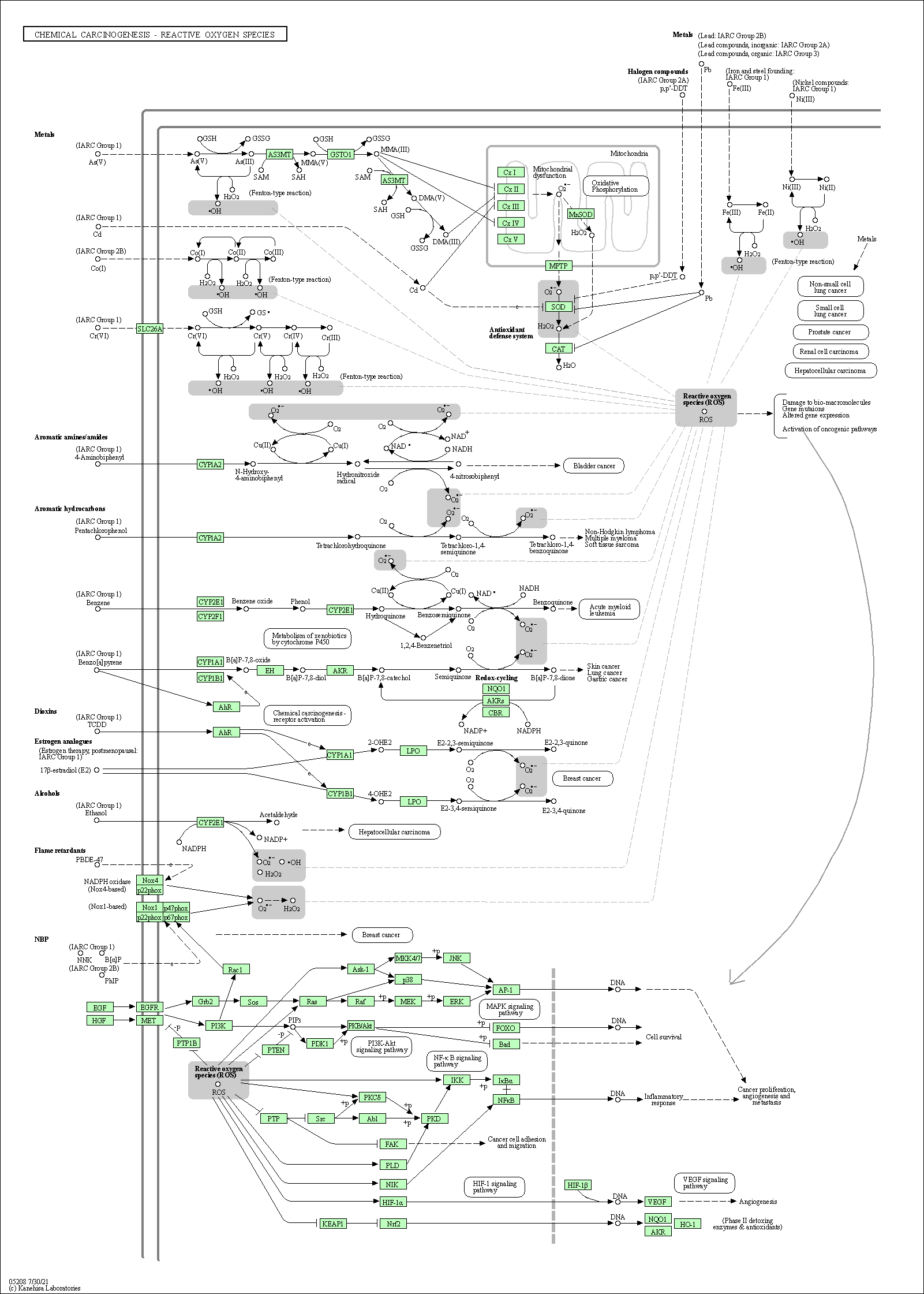
|
|||||||
| PD-L1 expression and PD-1 checkpoint pathway in cancer | hsa05235 |
Pathway Map 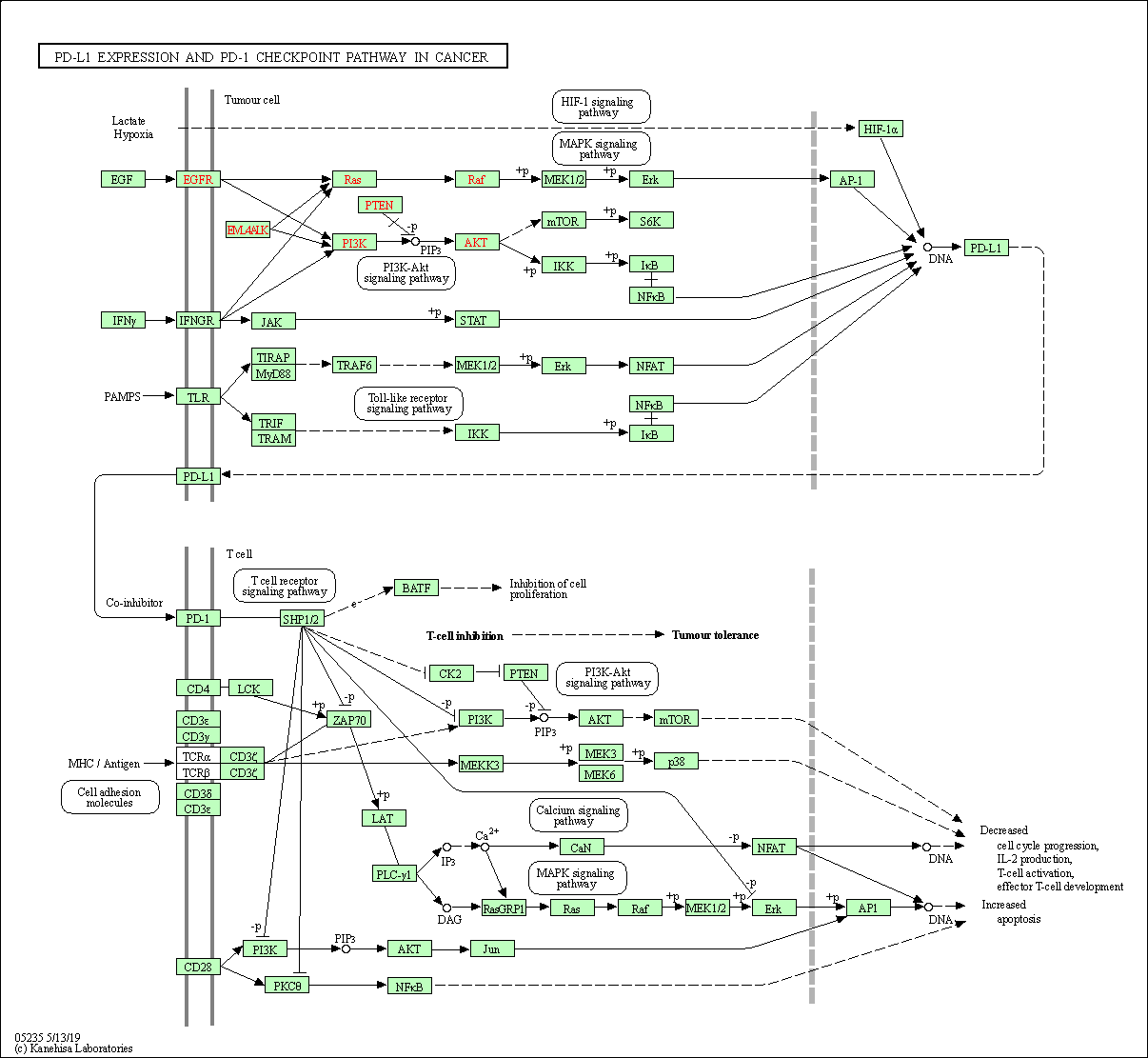
|
|||||||
| Pancreatic cancer | hsa05212 |
Pathway Map 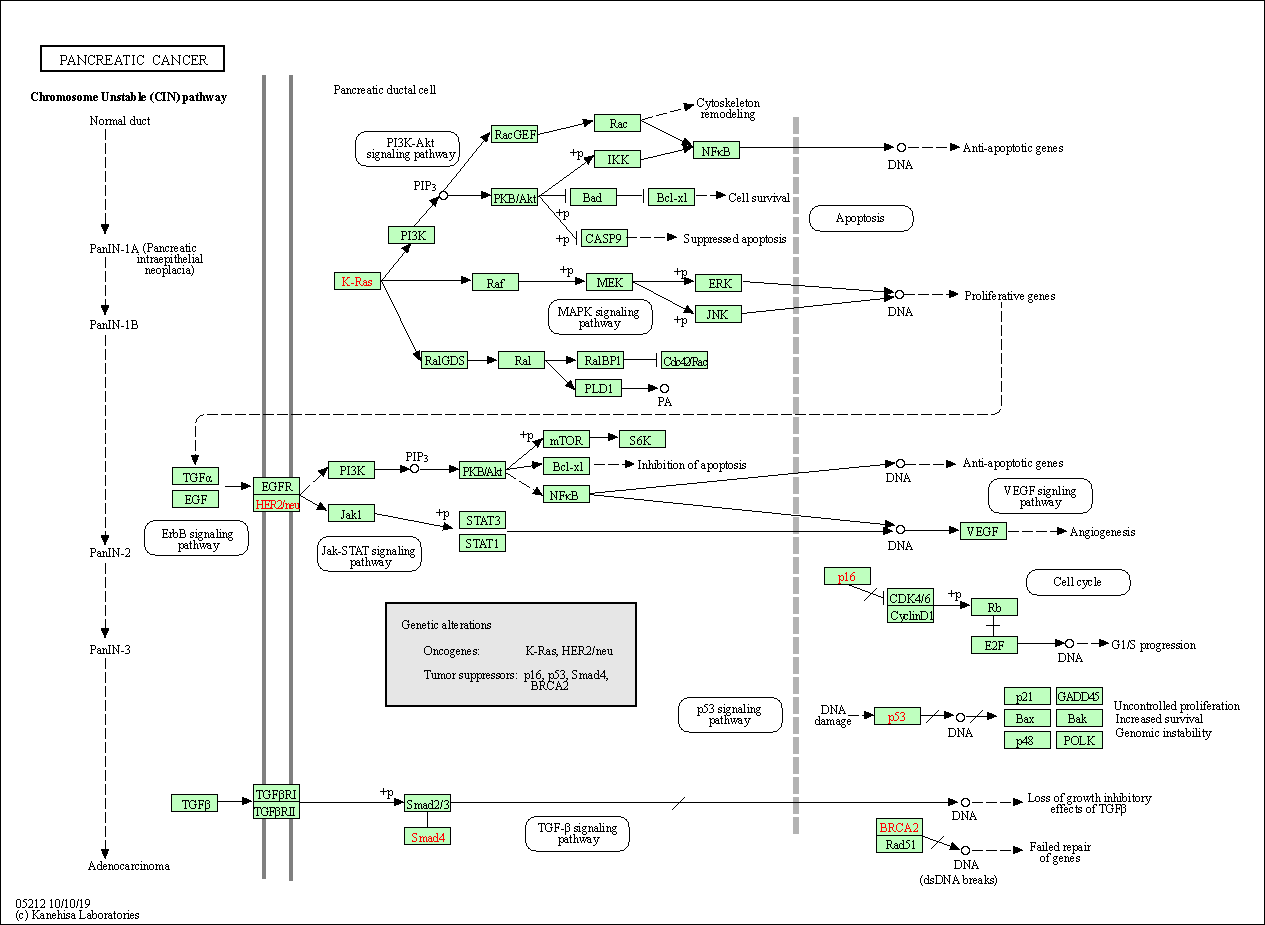
|
|||||||
| Prostate cancer | hsa05215 |
Pathway Map 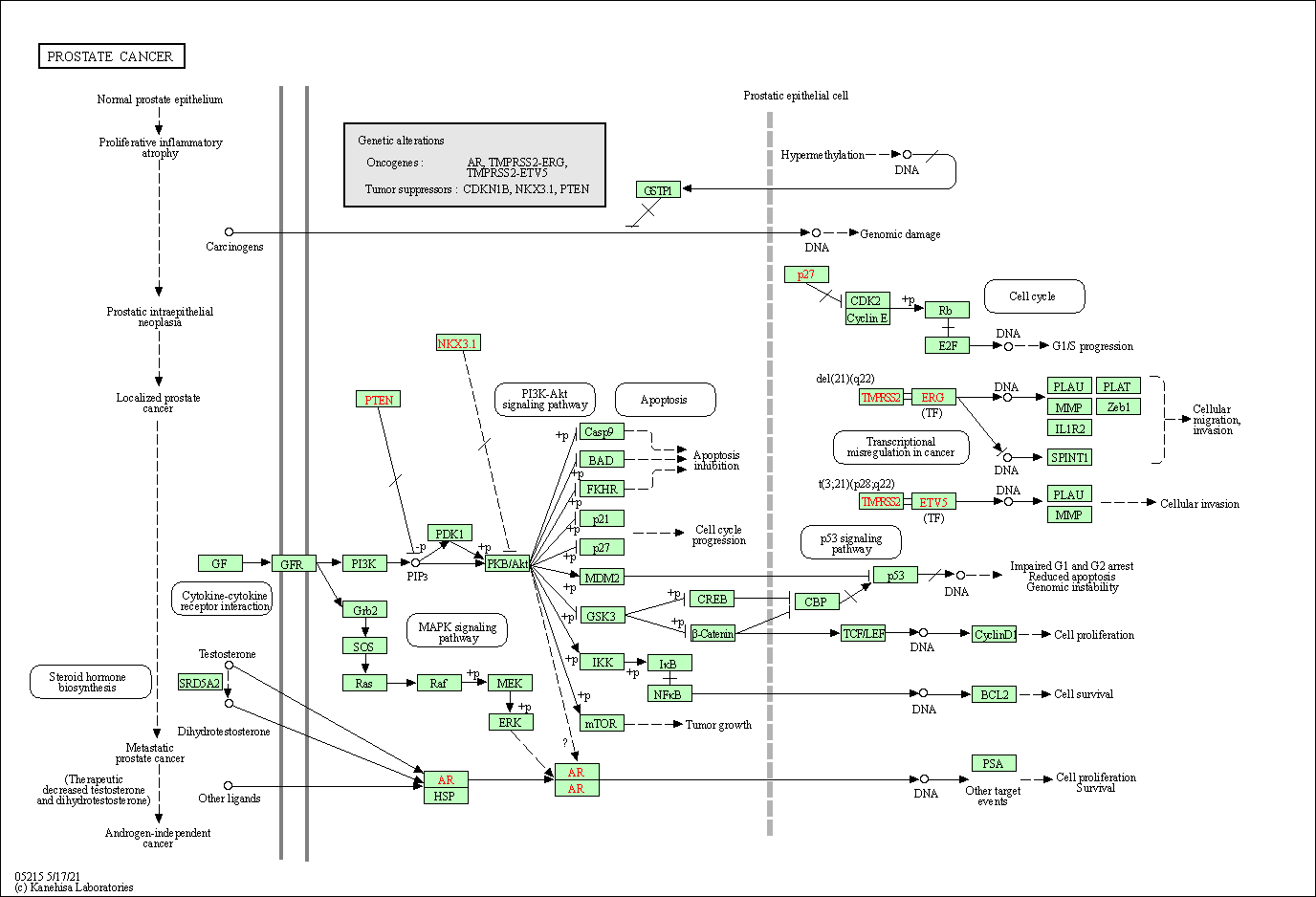
|
|||||||
| Chronic myeloid leukemia | hsa05220 |
Pathway Map 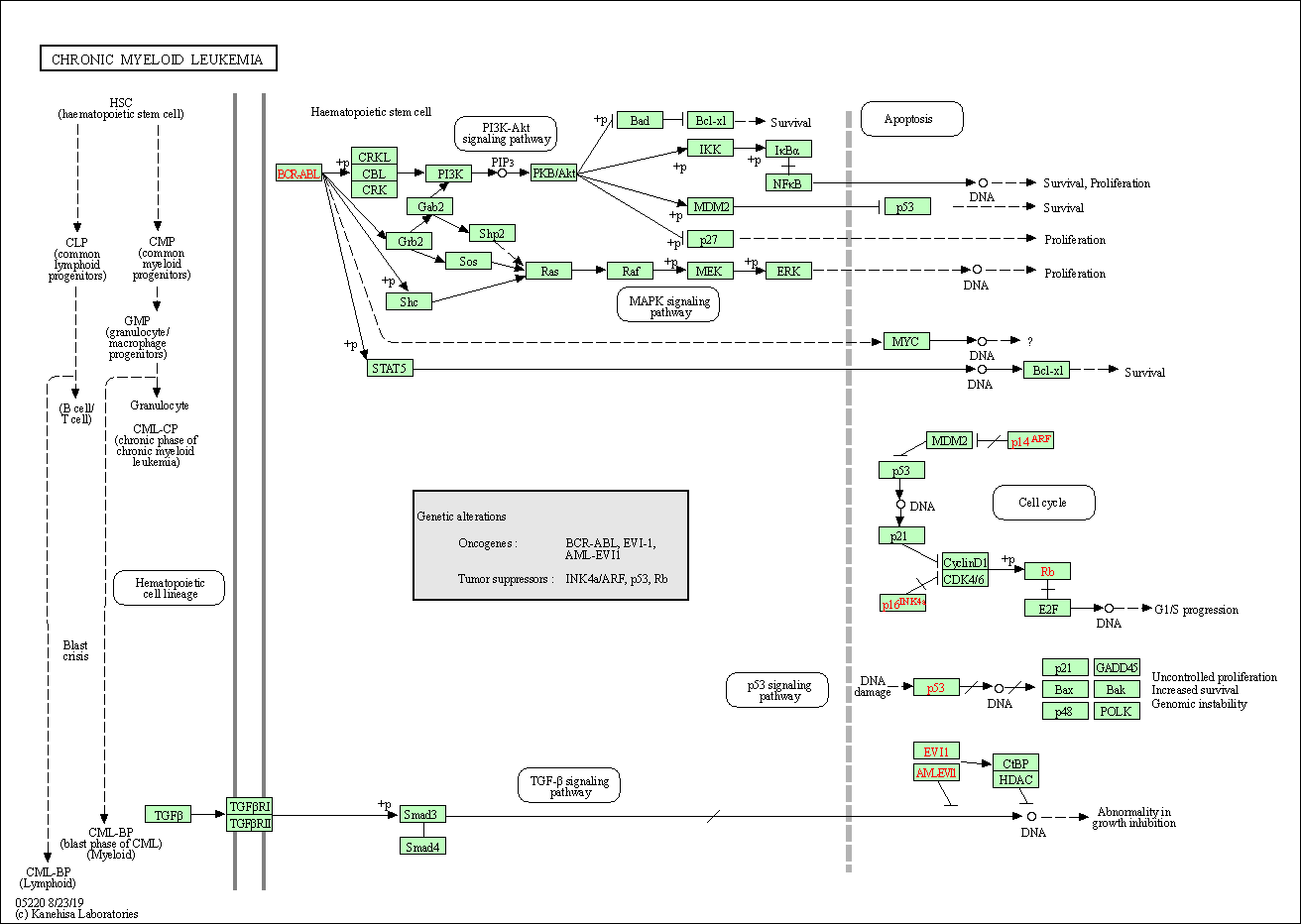
|
|||||||
| Acute myeloid leukemia | hsa05221 |
Pathway Map 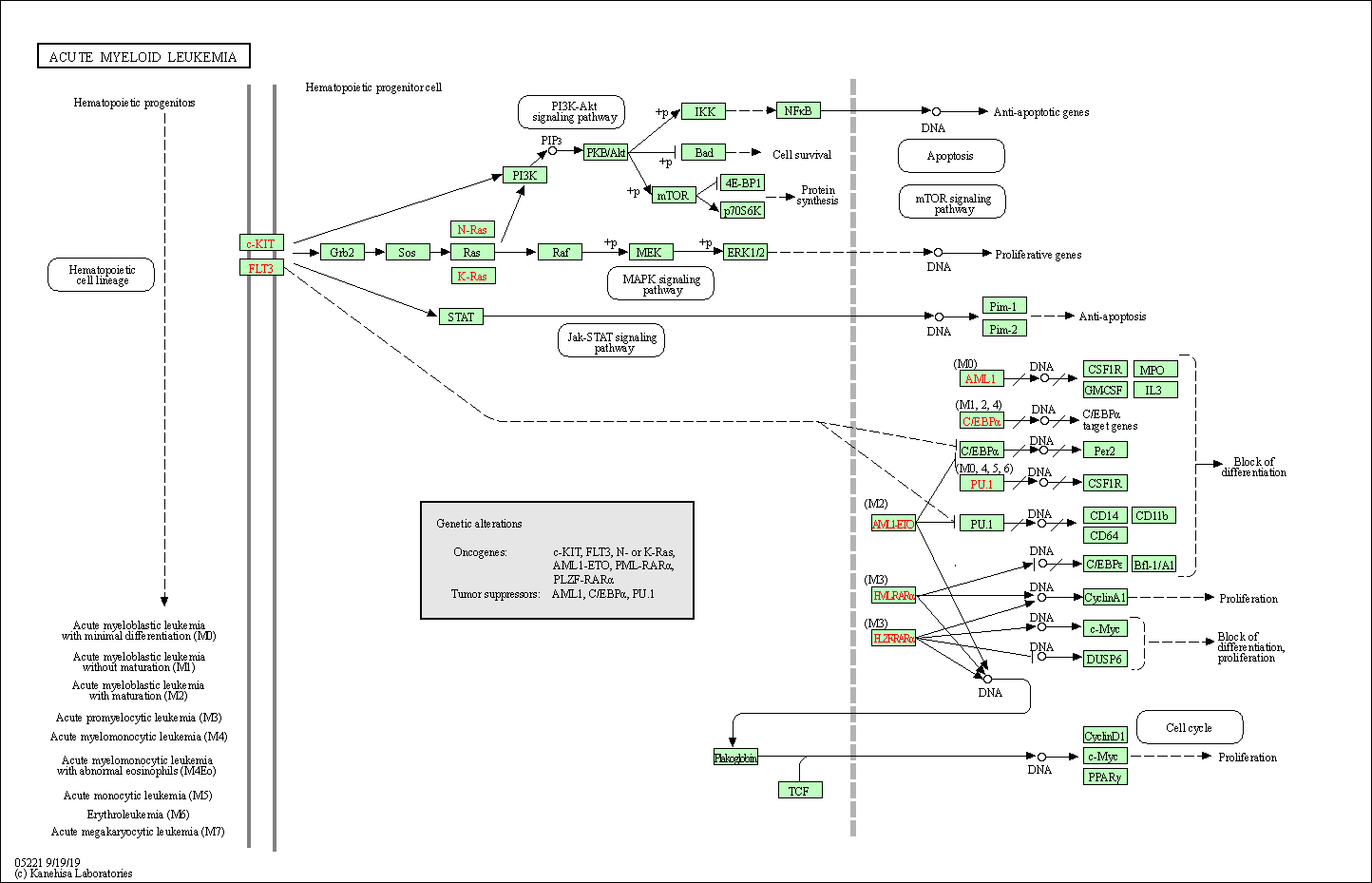
|
|||||||
| Small cell lung cancer | hsa05222 |
Pathway Map 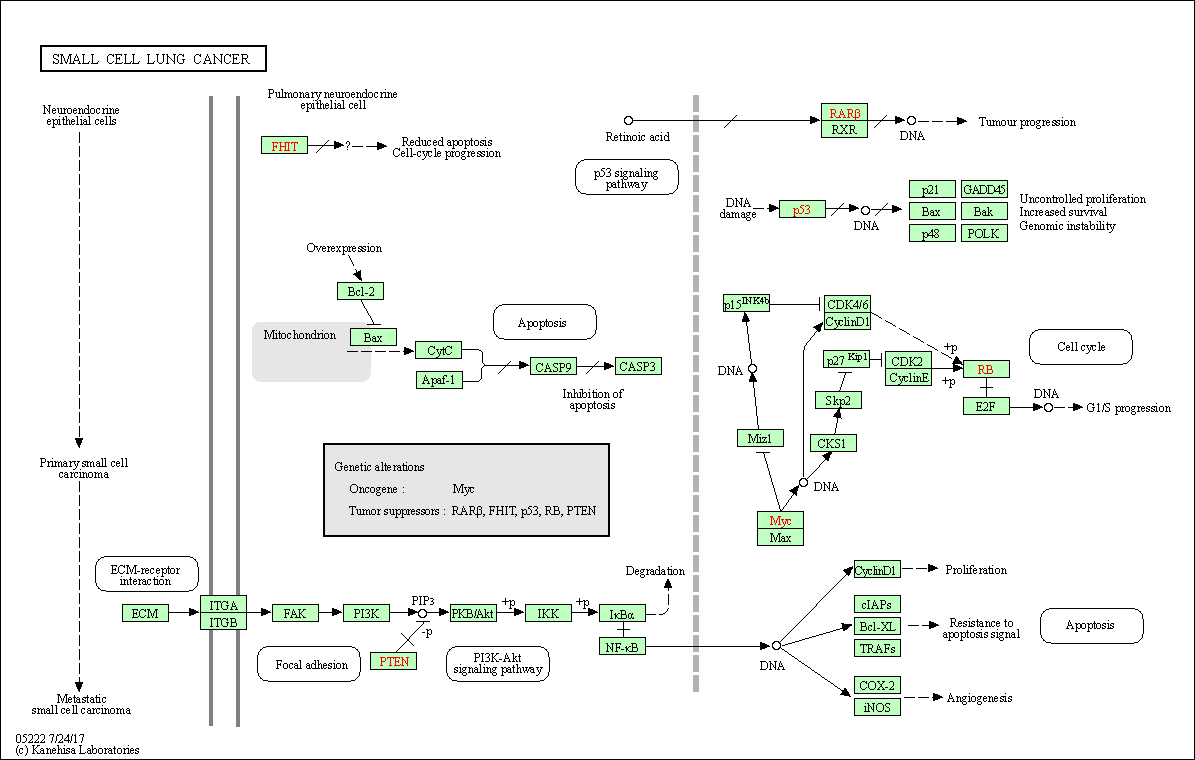
|
|||||||
| Diabetic cardiomyopathy | hsa05415 |
Pathway Map 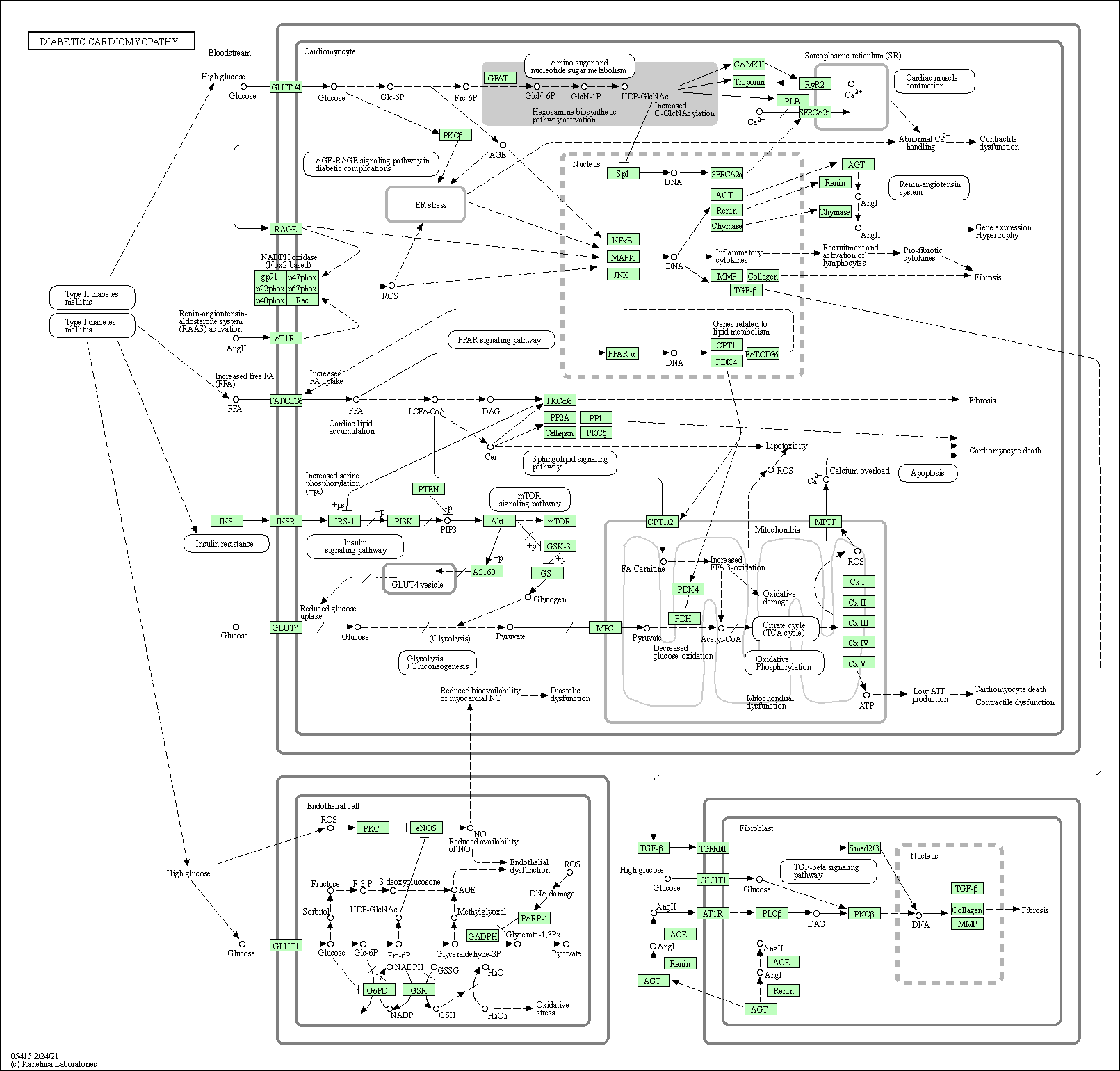
|
|||||||
| Lipid and atherosclerosis | hsa05417 |
Pathway Map 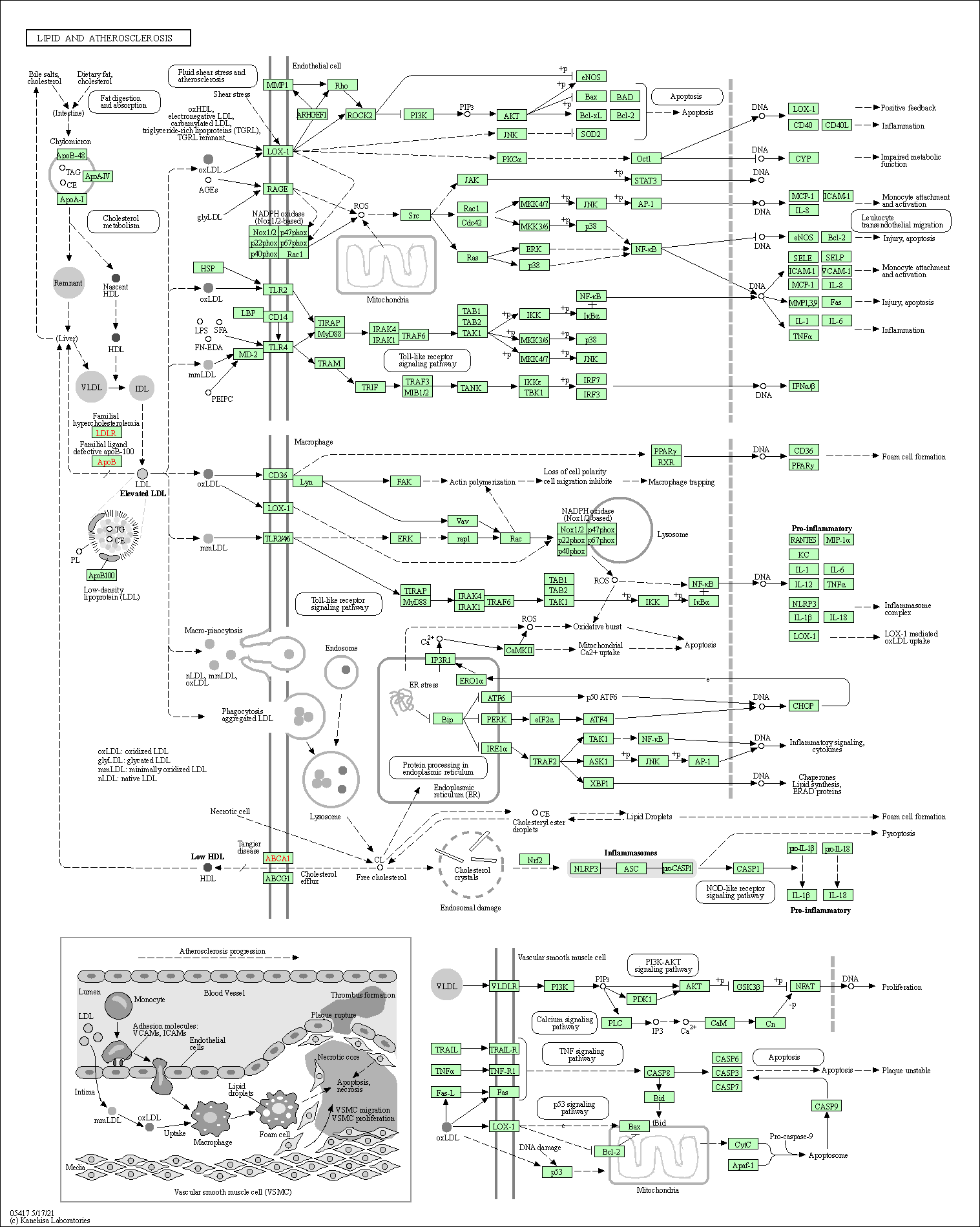
|
|||||||
| Fluid shear stress and atherosclerosis | hsa05418 |
Pathway Map 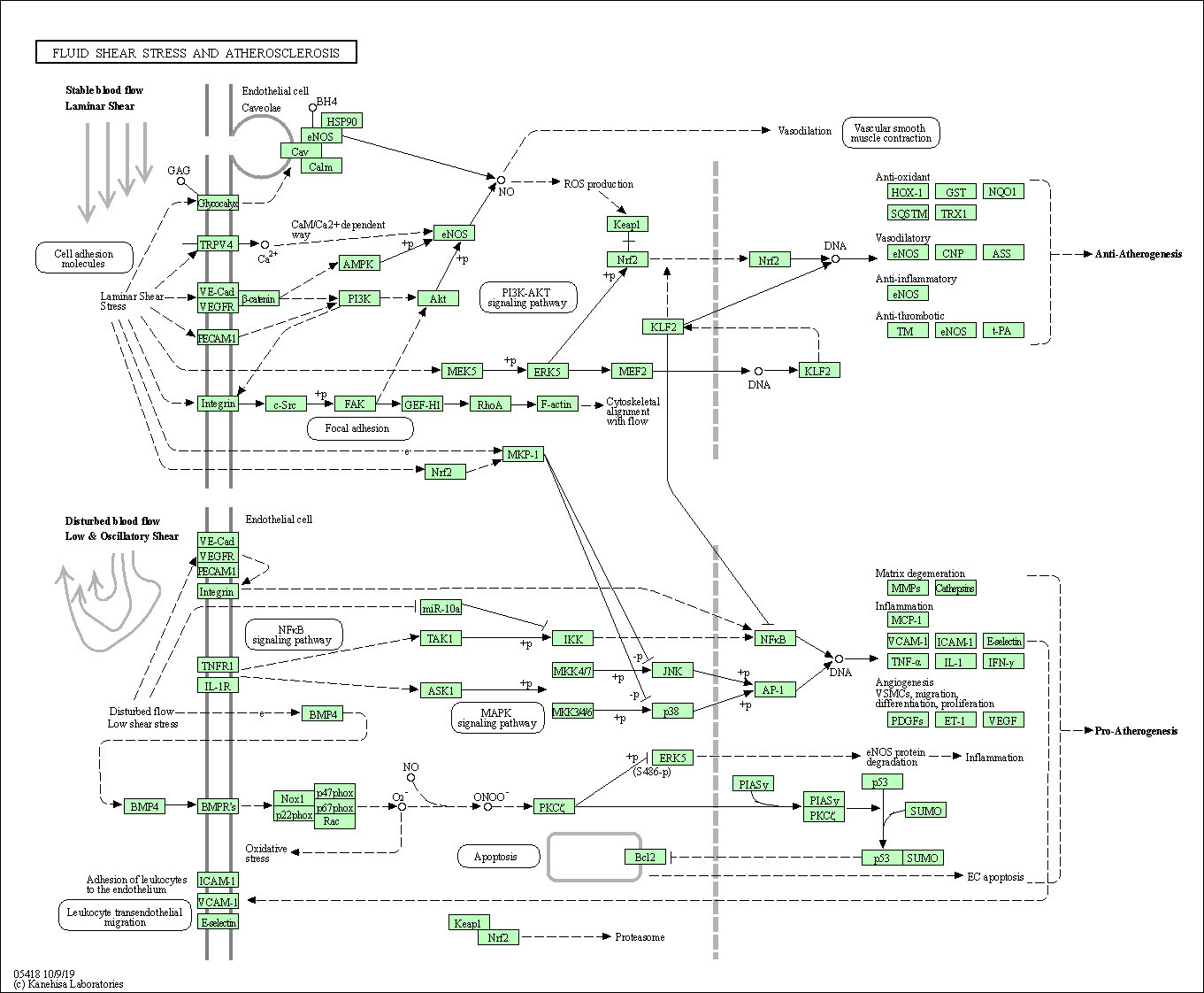
|
|||||||
| Apoptosis | hsa04210 |
Pathway Map 
|
|||||||
| Cellular senescence | hsa04218 |
Pathway Map 
|
|||||||
| Osteoclast differentiation | hsa04380 |
Pathway Map 
|
|||||||
| Insulin resistance | hsa04931 |
Pathway Map 
|
|||||||
| Non-alcoholic fatty liver disease | hsa04932 |
Pathway Map 
|
|||||||
| AGE-RAGE signaling pathway in diabetic complications | hsa04933 |
Pathway Map 
|
|||||||
| Alcoholic liver disease | hsa04936 |
Pathway Map 
|
|||||||
| Prolactin signaling pathway | hsa04917 |
Pathway Map 
|
|||||||
| Adipocytokine signaling pathway | hsa04920 |
Pathway Map 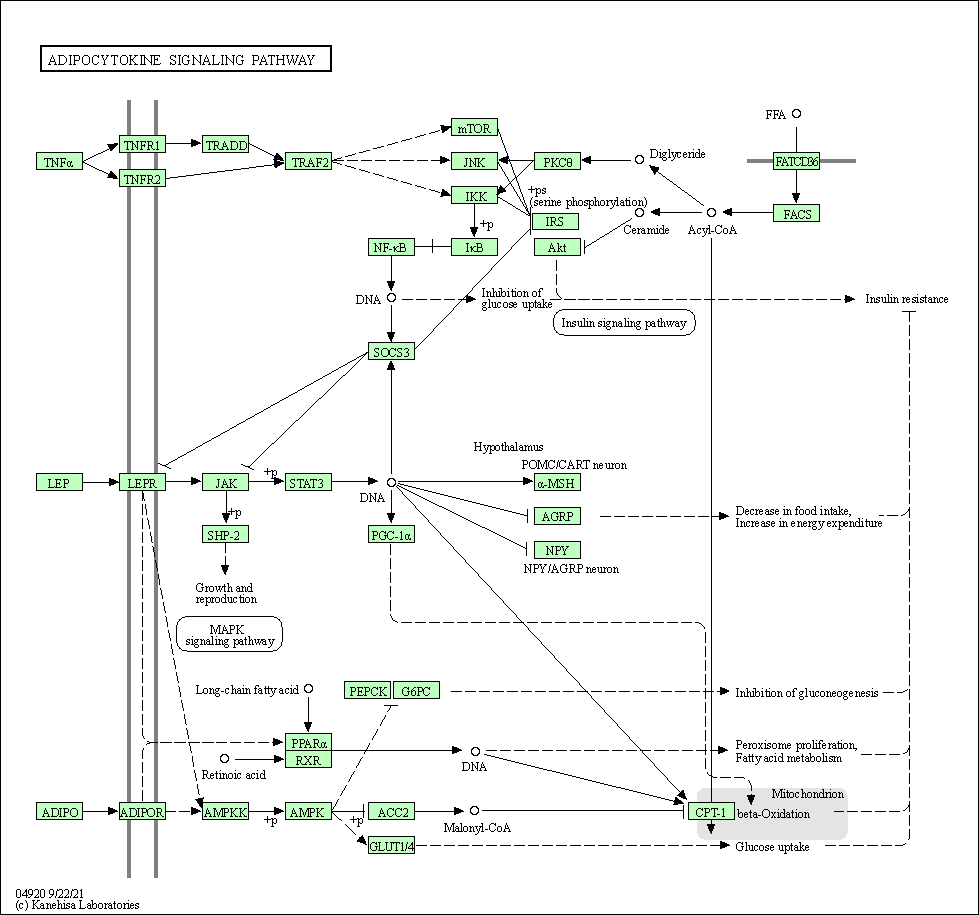
|
|||||||
| Relaxin signaling pathway | hsa04926 |
Pathway Map 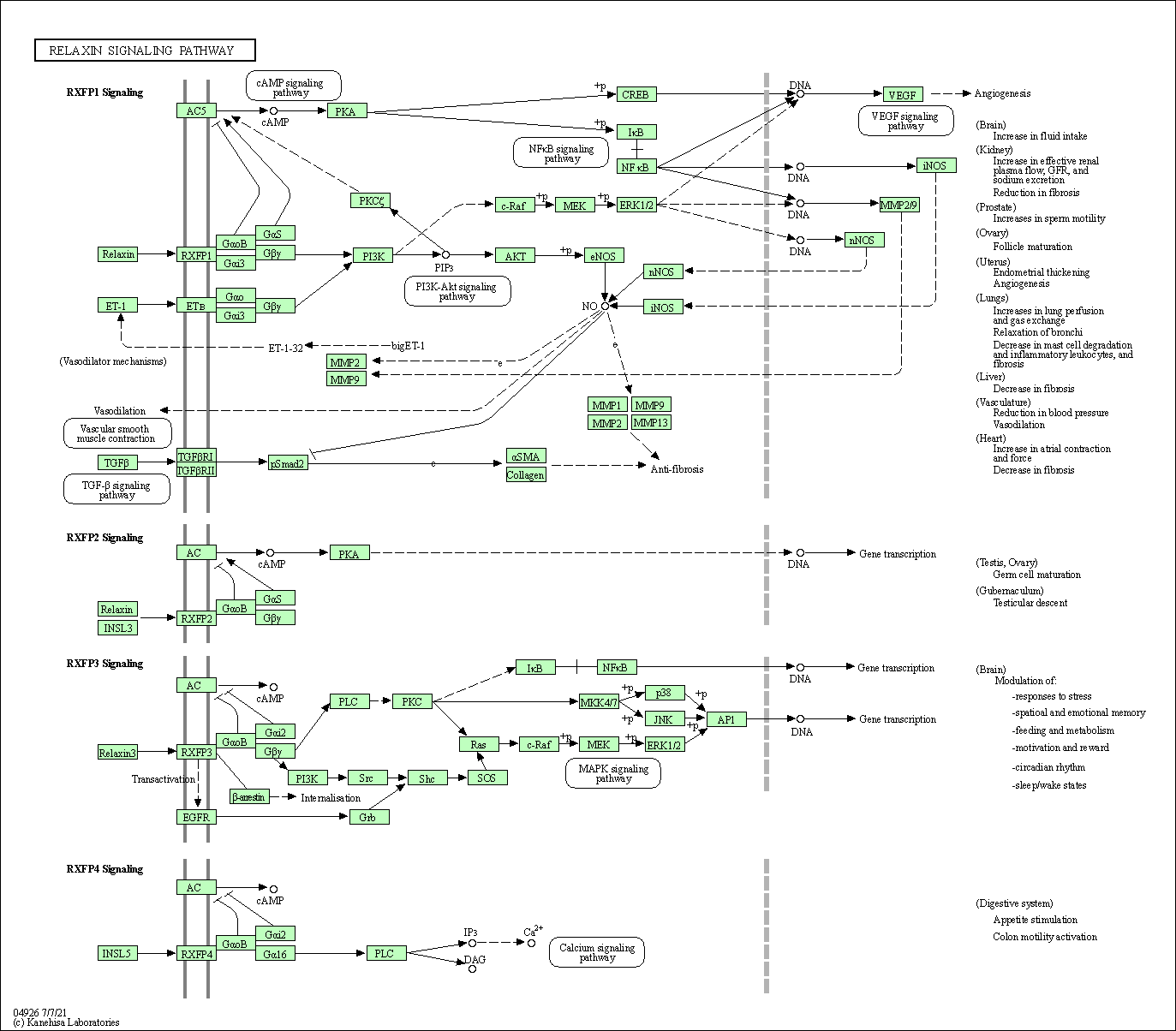
|
|||||||
| Inflammatory bowel disease | hsa05321 |
Pathway Map 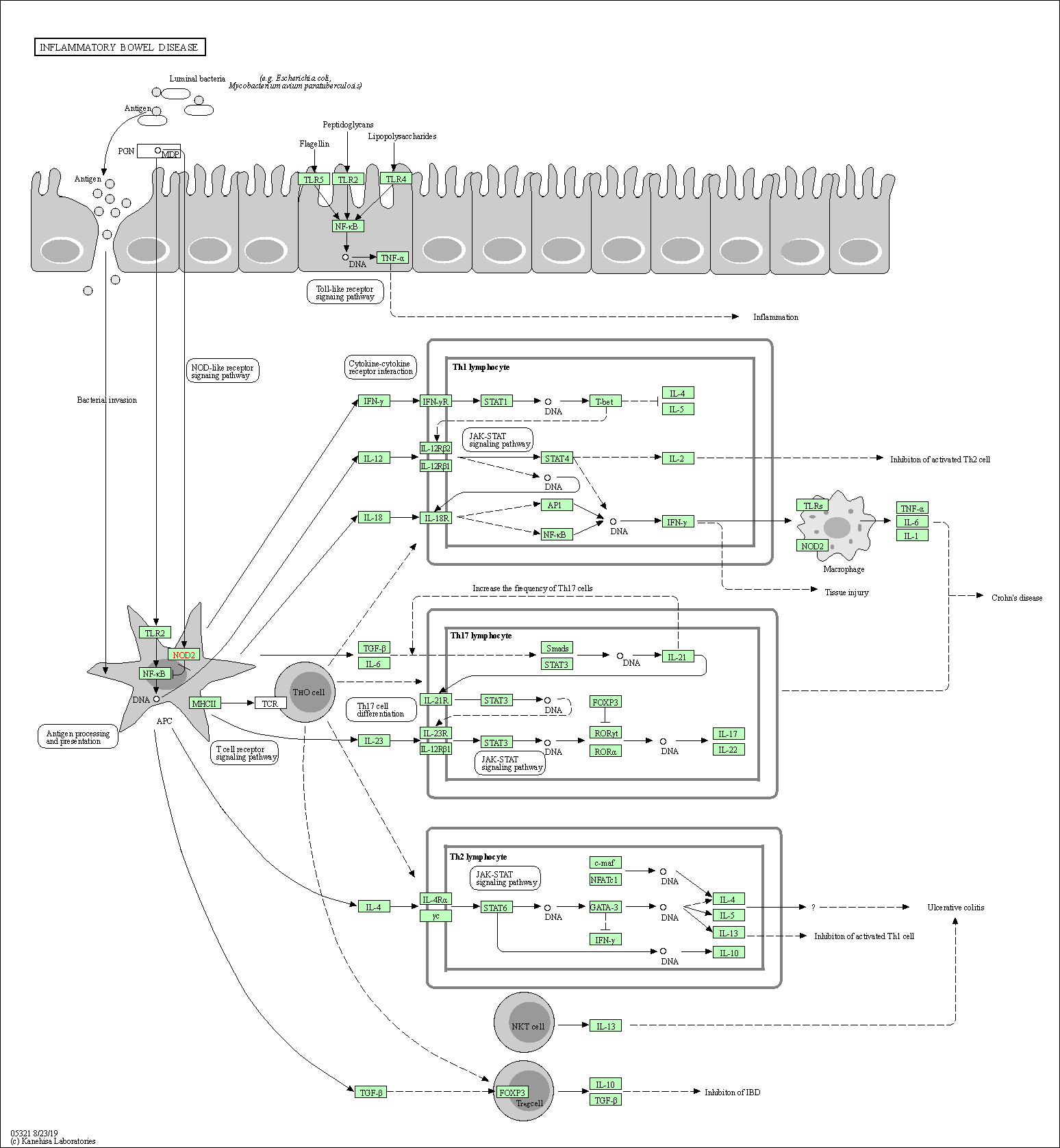
|
|||||||
| Neurotrophin signaling pathway | hsa04722 |
Pathway Map 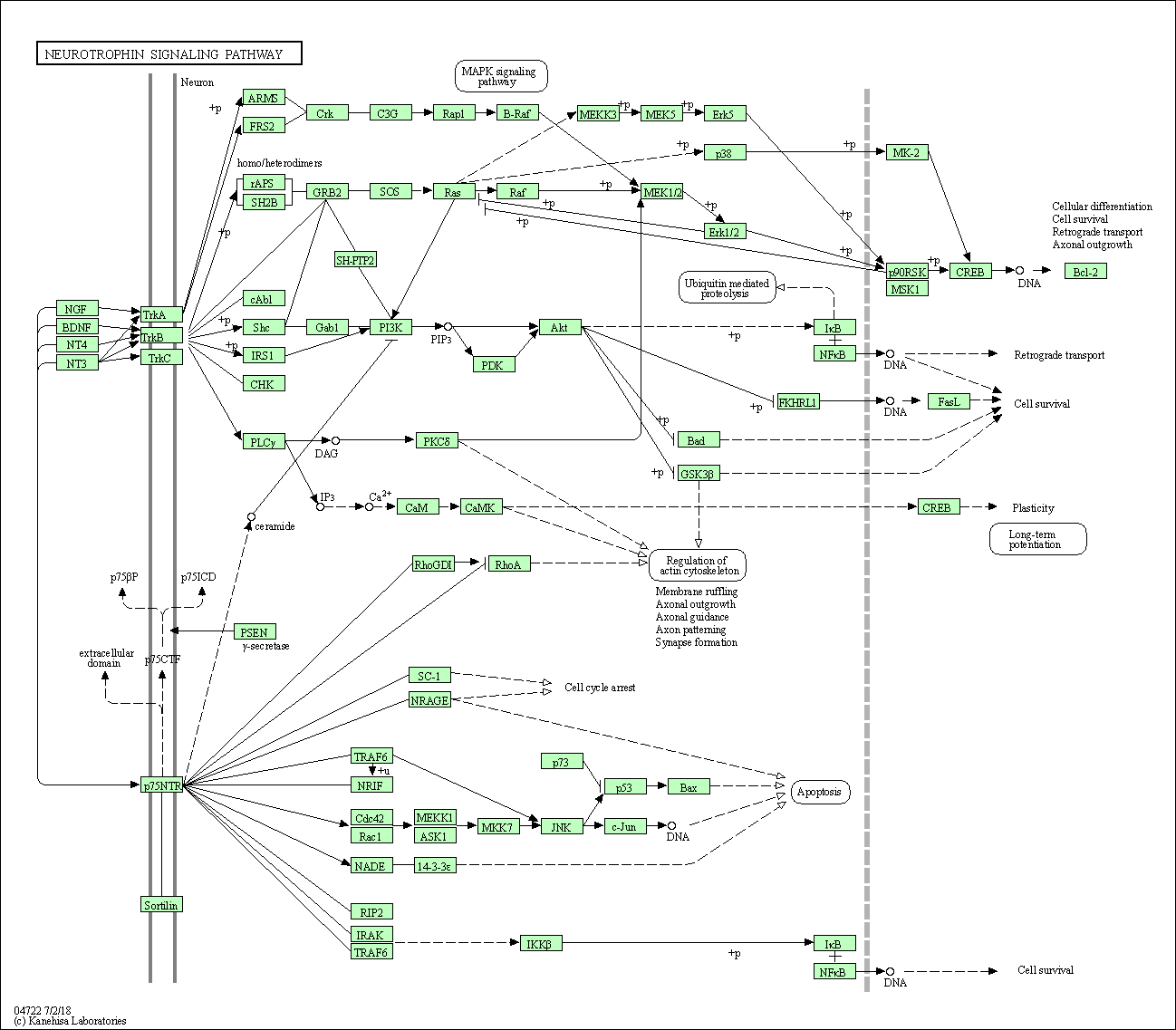
|
|||||||
| Alzheimer disease | hsa05010 |
Pathway Map 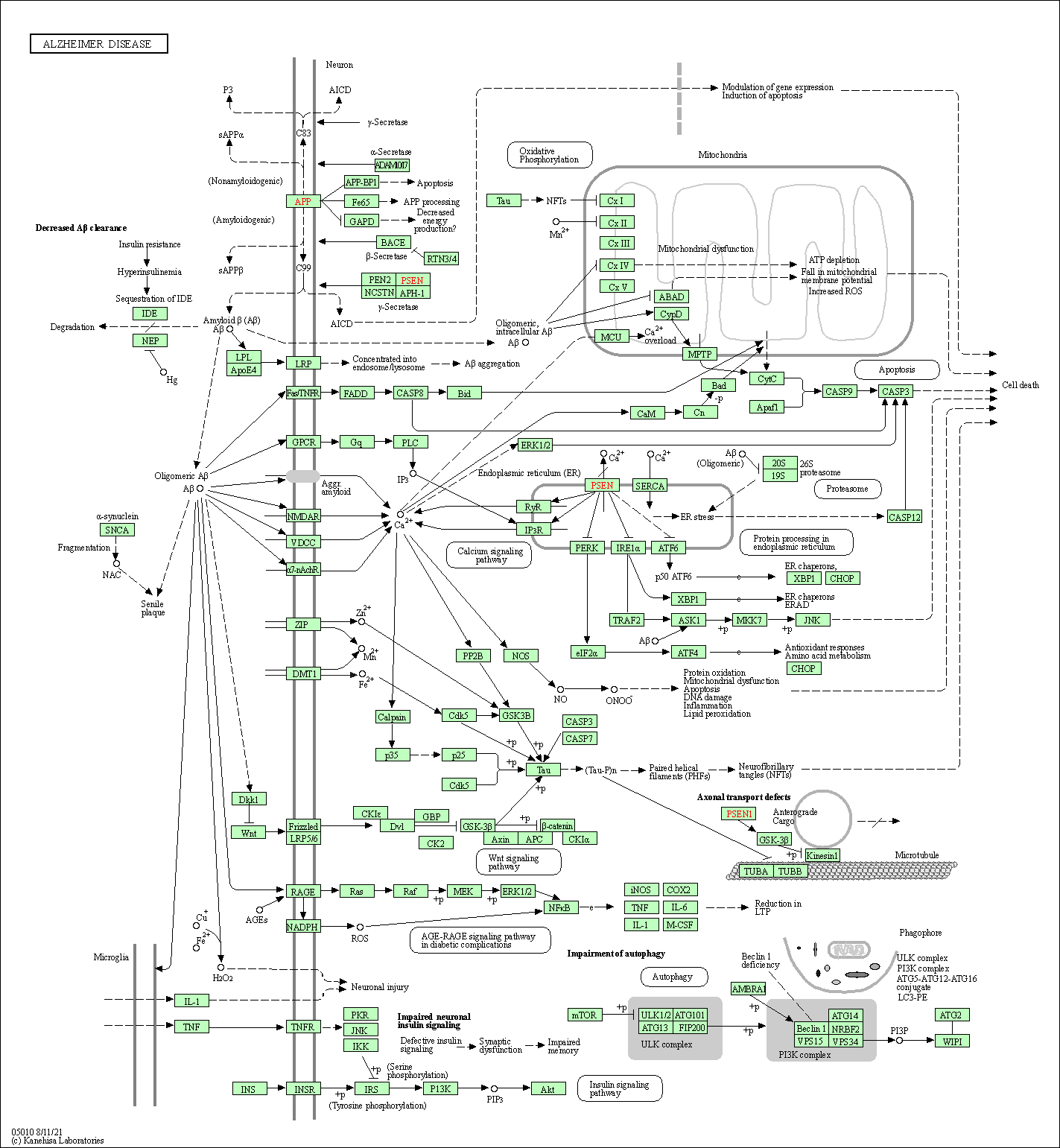
|
|||||||
| Pathways of neurodegeneration - multiple diseases | hsa05022 |
Pathway Map 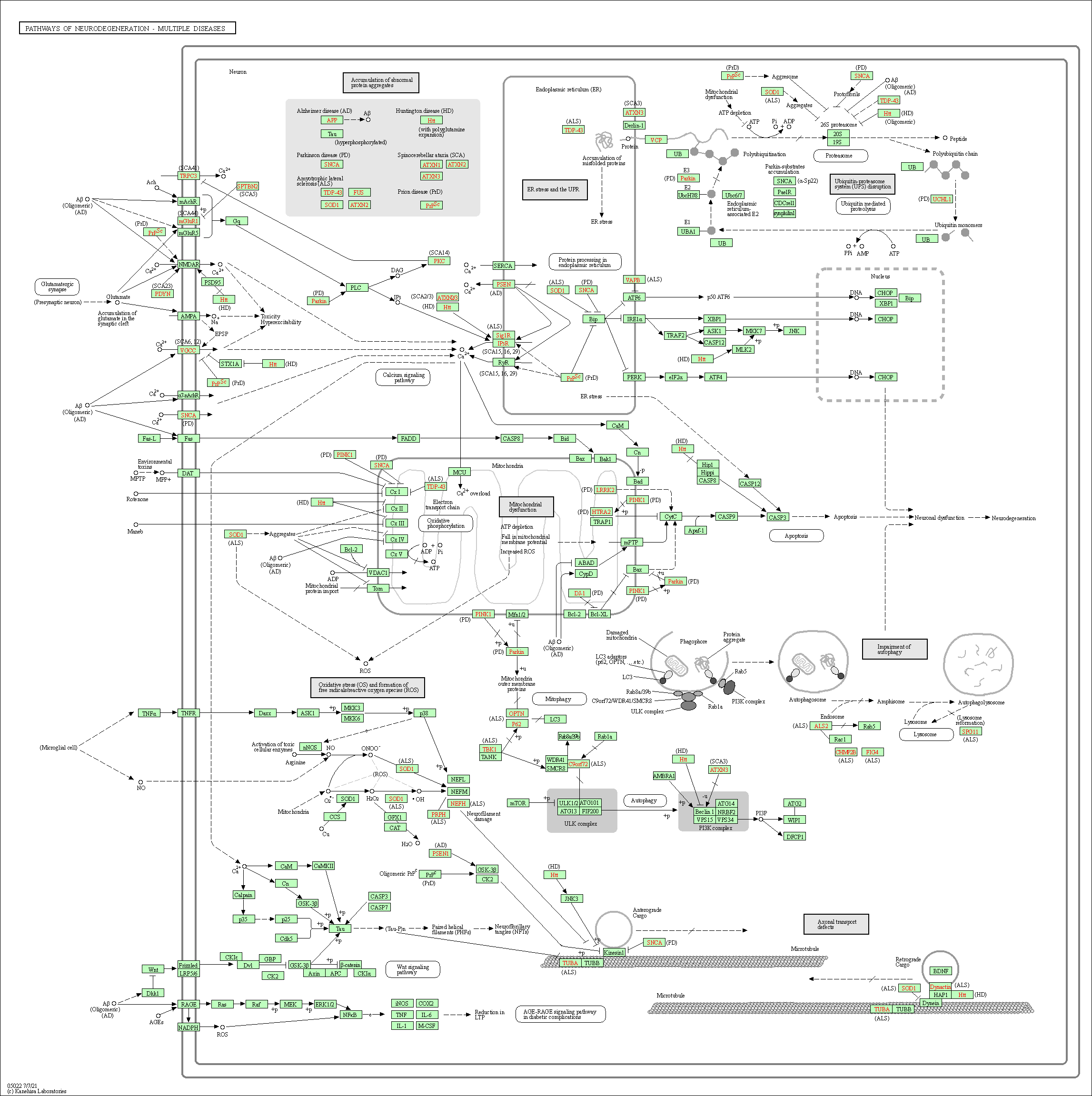
|
|||||||
| MAPK signaling pathway | hsa04010 |
Pathway Map 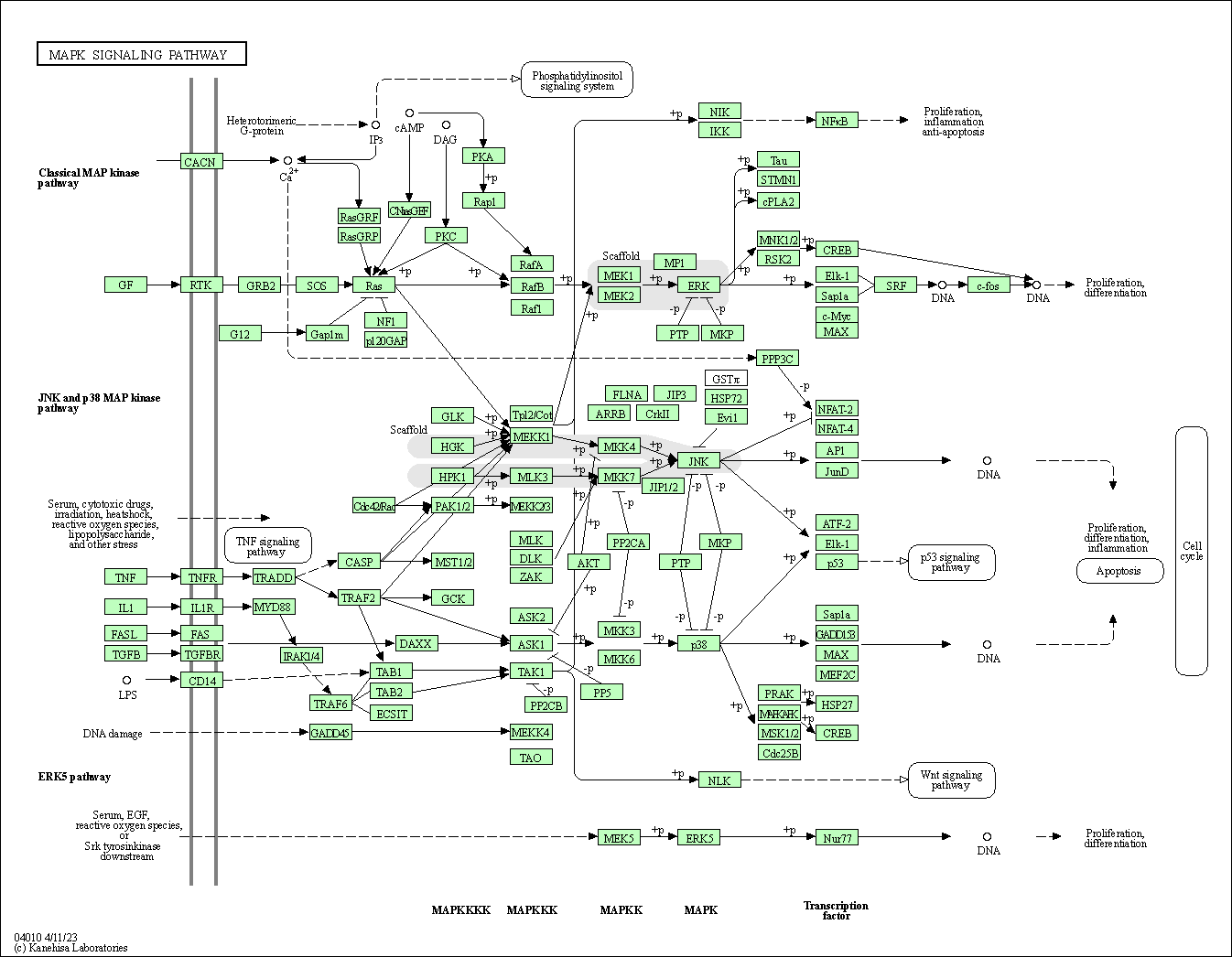
|
|||||||
| Ras signaling pathway | hsa04014 |
Pathway Map 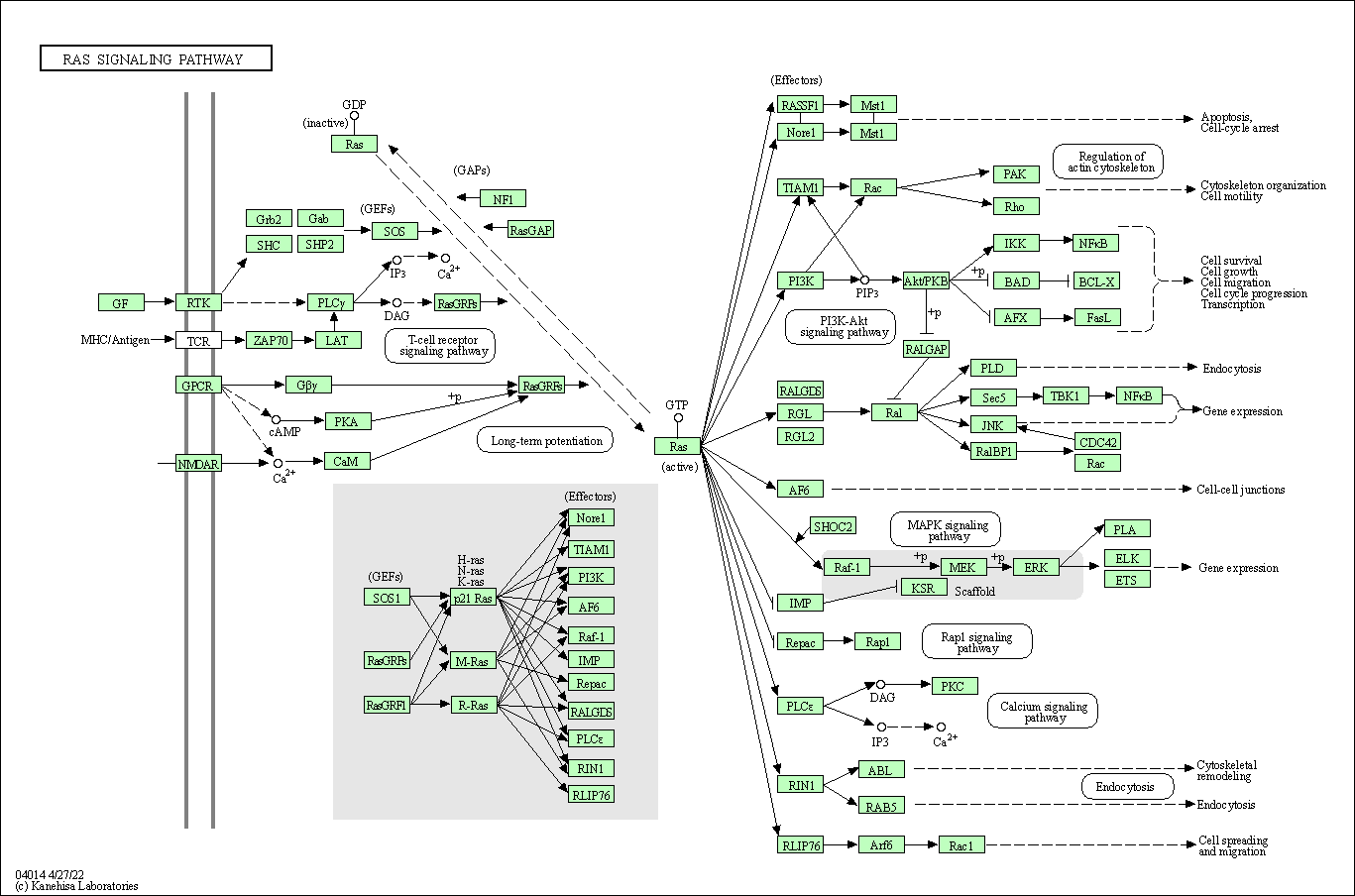
|
|||||||
| cAMP signaling pathway | hsa04024 |
Pathway Map 
|
|||||||
| NF-kappa B signaling pathway | hsa04064 |
Pathway Map 
|
|||||||
| HIF-1 signaling pathway | hsa04066 |
Pathway Map 
|
|||||||
| Sphingolipid signaling pathway | hsa04071 |
Pathway Map 
|
|||||||
| PI3K-Akt signaling pathway | hsa04151 |
Pathway Map 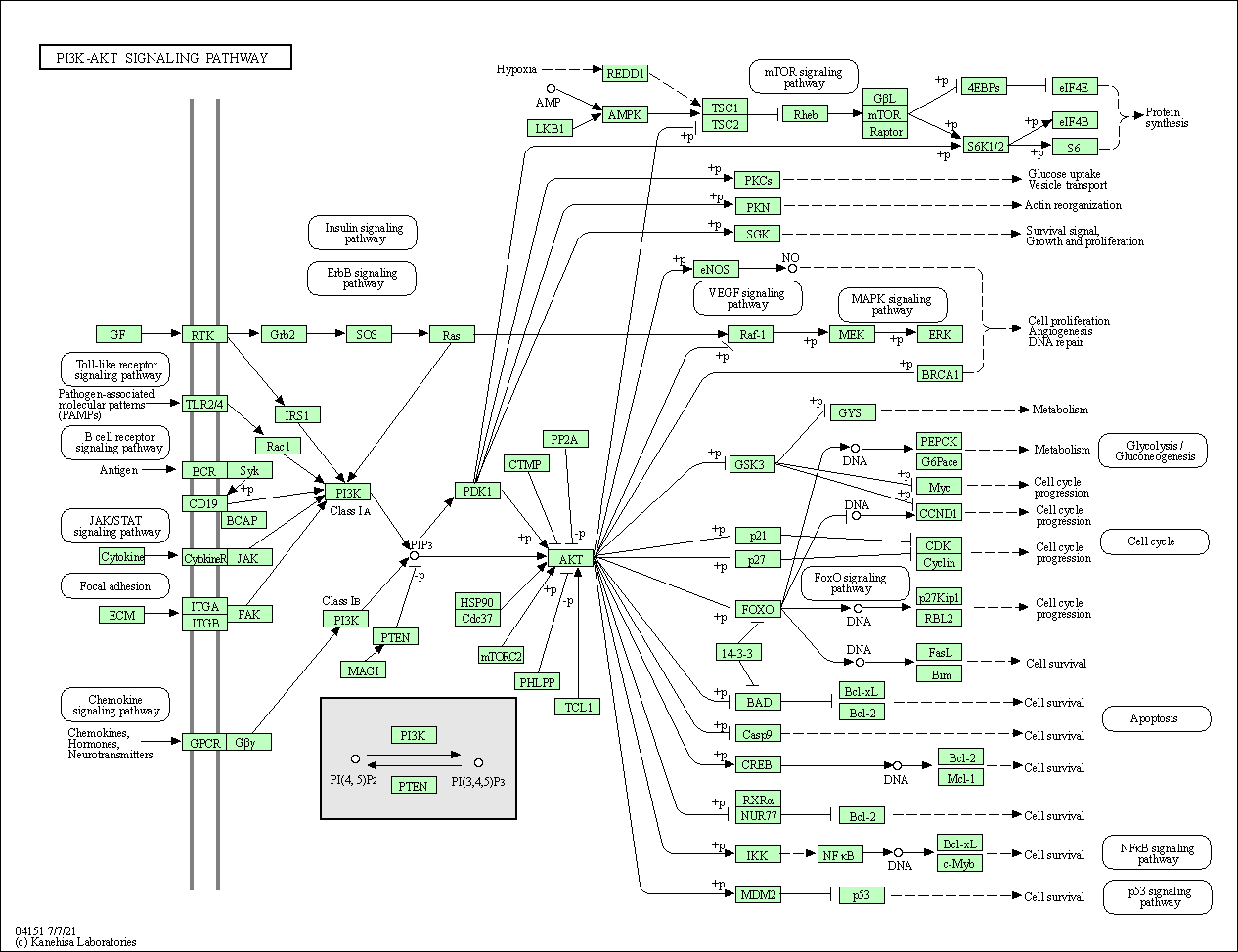
|
|||||||
| TNF signaling pathway | hsa04668 |
Pathway Map 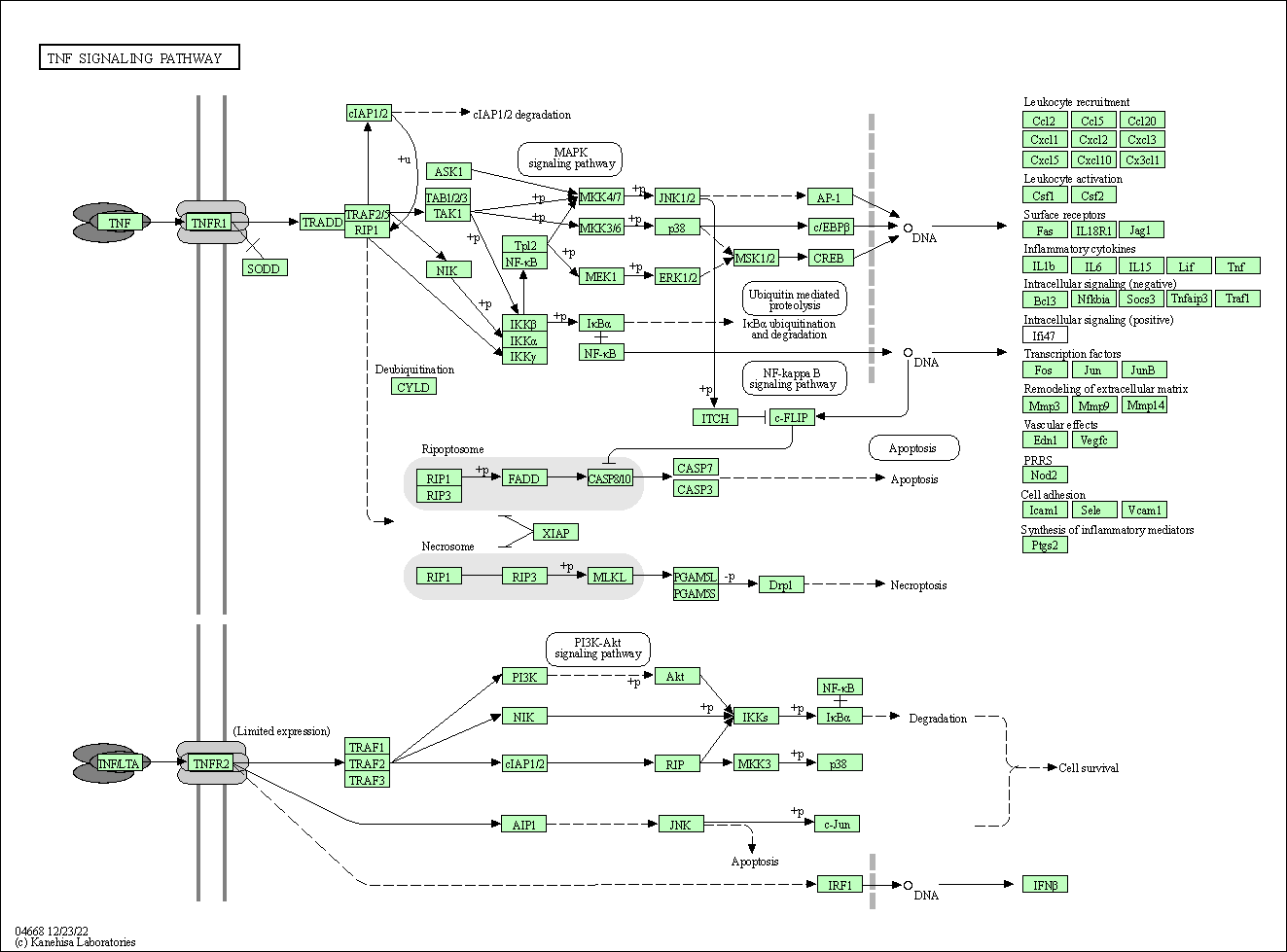
|
|||||||
| Cocaine addiction | hsa05030 |
Pathway Map 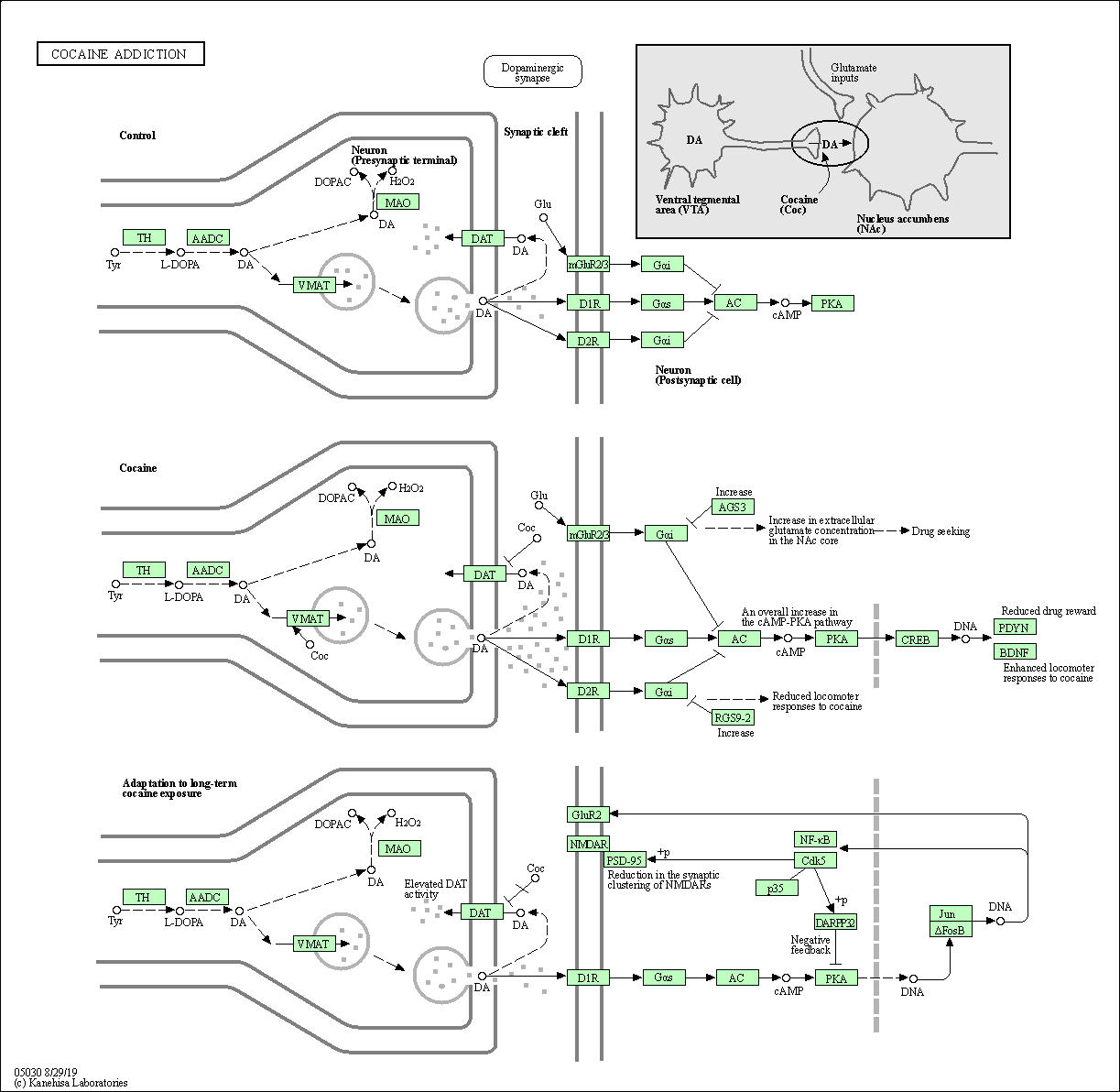
|
|||||||
| Mitophagy - animal | hsa04137 |
Pathway Map 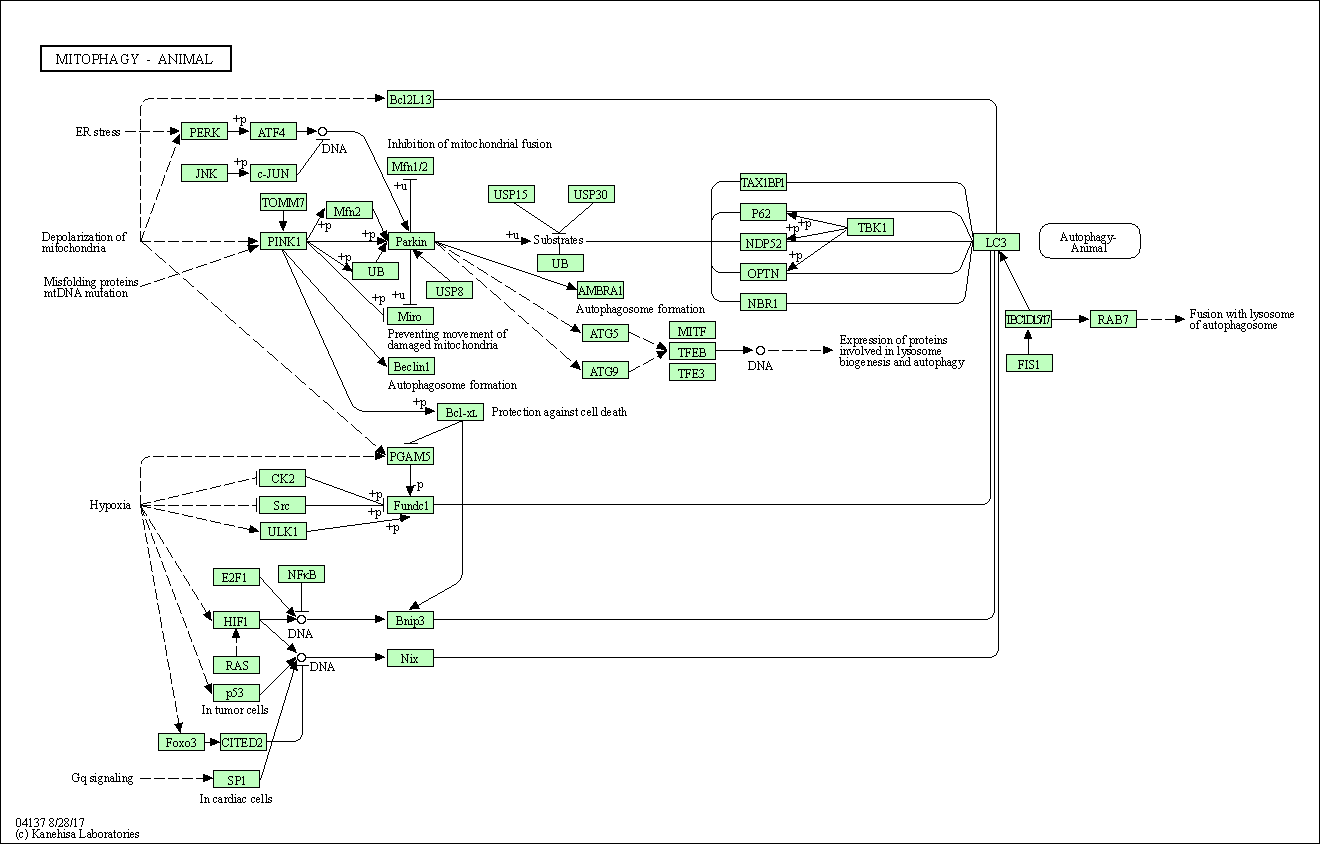
|
|||||||
| 3D Structure |
|
||||||||
Full List of Virus RNA Interacting with This Protien
| Virus Name | Binding Region | RNA Info | Detection Method | Infection Cell | Cell ID | Cell Originated Tissue | Infection Time | Interaction Score 1 | Interaction Score 2 |
|---|---|---|---|---|---|---|---|---|---|
| Hepatitis E virus | 3'UTR | RNA Info | RNA-protein interaction detection (RaPID) assay coupled to liquid chromatography with tandem mass spectrometry (LC-MS/MS) | HEK293 Cells (Human embryonic kidney cell); Mesc cells (Embryonic stem cell) | . | Kidney | . | . | . |
| Influenza A virus (strain California/7/2004 (H3N2)) | 5'UTR - 3'UTR | RNA Info | VIRal Cross-Linking And Solid-phase Purification (VIR-CLASP) | BHK-21 Cells (Small hamster kidney fibroblast) | . | Kidney | 1 h | P = 0.000128453945849677 | . |
| Chikungunya virus (strain 181/25) | 5'UTR - 3'UTR | RNA Info | VIRal Cross-Linking And Solid-phase Purification (VIR-CLASP) | BHK-21 Cells (Small hamster kidney fibroblast) | . | Kidney | 1 h | P = 9.30254812904301e-05 | . |
| Chikungunya virus (strain 181/25) | 5'UTR - 3'UTR | RNA Info | VIRal Cross-Linking And Solid-phase Purification (VIR-CLASP) | BHK-21 Cells (Small hamster kidney fibroblast) | . | Kidney | 3 h | P = 0.000249176554464078 | . |
| Chikungunya virus (strain 181/25) | 5'UTR - 3'UTR | RNA Info | VIRal Cross-Linking And Solid-phase Purification (VIR-CLASP) | BHK-21 Cells (Small hamster kidney fibroblast) | . | Kidney | 0.2 h | P = 0.000125809176284766 | . |
Potential Drug(s) that Targets This Protein
Protein Sequence Information
|
MDELFPLIFPAEPAQASGPYVEIIEQPKQRGMRFRYKCEGRSAGSIPGERSTDTTKTHPTIKINGYTGPGTVRISLVTKDPPHRPHPHELVGKDCRDGFYEAELCPDRCIHSFQNLGIQCVKKRDLEQAISQRIQTNNNPFQVPIEEQRGDYDLNAVRLCFQVTVRDPSGRPLRLPPVLSHPIFDNRAPNTAELKICRVNRNSGSCLGGDEIFLLCDKVQKEDIEVYFTGPGWEARGSFSQADVHRQVAIVFRTPPYADPSLQAPVRVSMQLRRPSDRELSEPMEFQYLPDTDDRHRIEEKRKRTYETFKSIMKKSPFSGPTDPRPPPRRIAVPSRSSASVPKPAPQPYPFTSSLSTINYDEFPTMVFPSGQISQASALAPAPPQVLPQAPAPAPAPAMVSALAQAPAPVPVLAPGPPQAVAPPAPKPTQAGEGTLSEALLQLQFDDEDLGALLGNSTDPAVFTDLASVDNSEFQQLLNQGIPVAPHTTEPMLMEYPEAITRLVTGAQRPPDPAPAPLGAPGLPNGLLSGDEDFSSIADMDFSALLSQISS
Click to Show/Hide
|
