Host Protein General Information
| Protein Name |
Isocitrate dehydrogenase [NADP] cytoplasmic
|
Gene Name |
IDH1
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Host Species |
Homo sapiens
|
Uniprot Entry Name |
IDHC_HUMAN
|
||||||
| Protein Families |
Isocitrate and isopropylmalate dehydrogenases family
|
||||||||
| EC Number |
1.1.1.42
|
||||||||
| Subcellular Location |
Cytoplasm; cytosol
|
||||||||
| External Link | |||||||||
| NCBI Gene ID | |||||||||
| Uniprot ID | |||||||||
| Ensembl ID | |||||||||
| HGNC ID | |||||||||
| Related KEGG Pathway | |||||||||
| Central carbon metabolism in cancer | hsa05230 |
Pathway Map 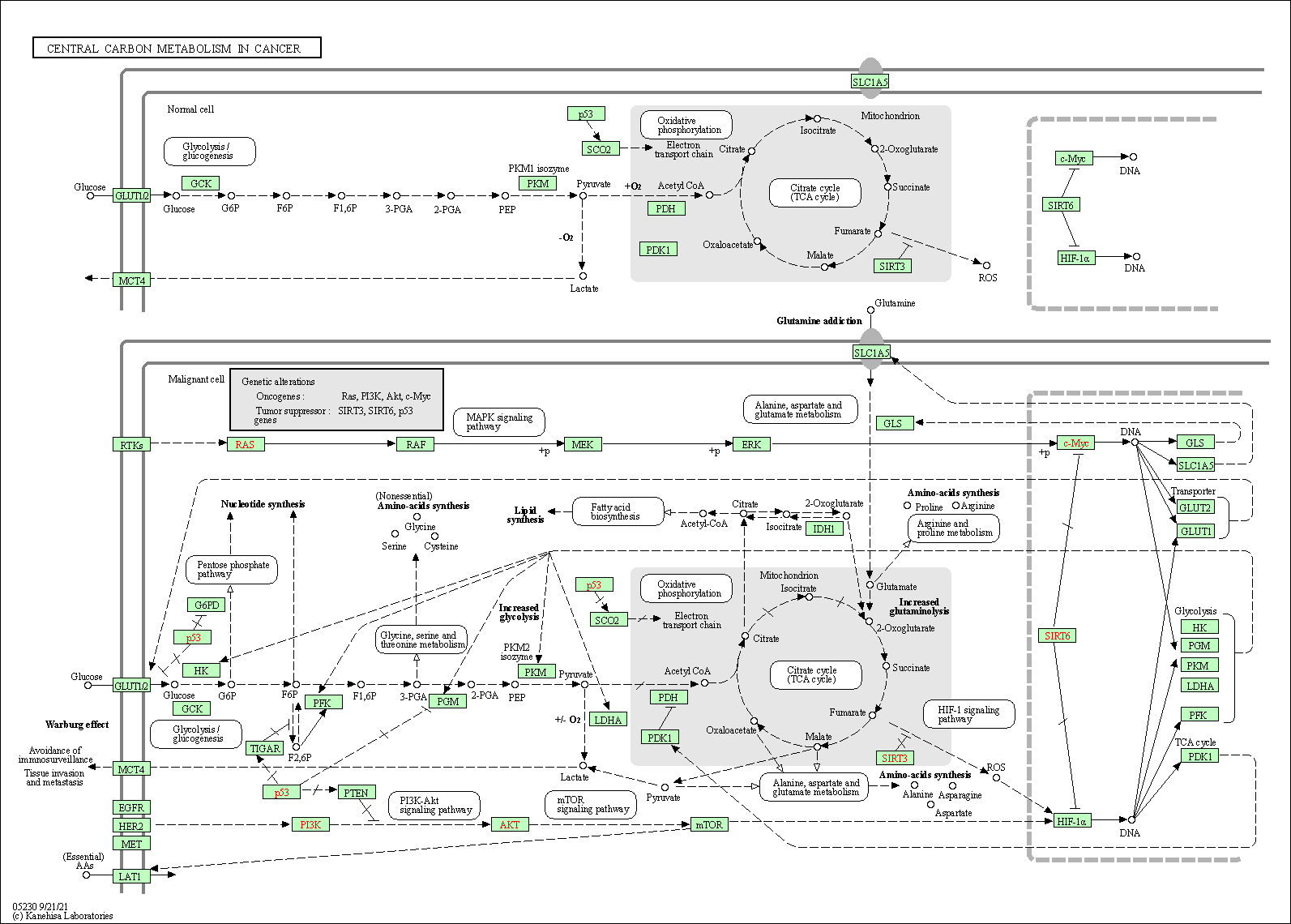
|
|||||||
| Citrate cycle (TCA cycle) | hsa00020 |
Pathway Map 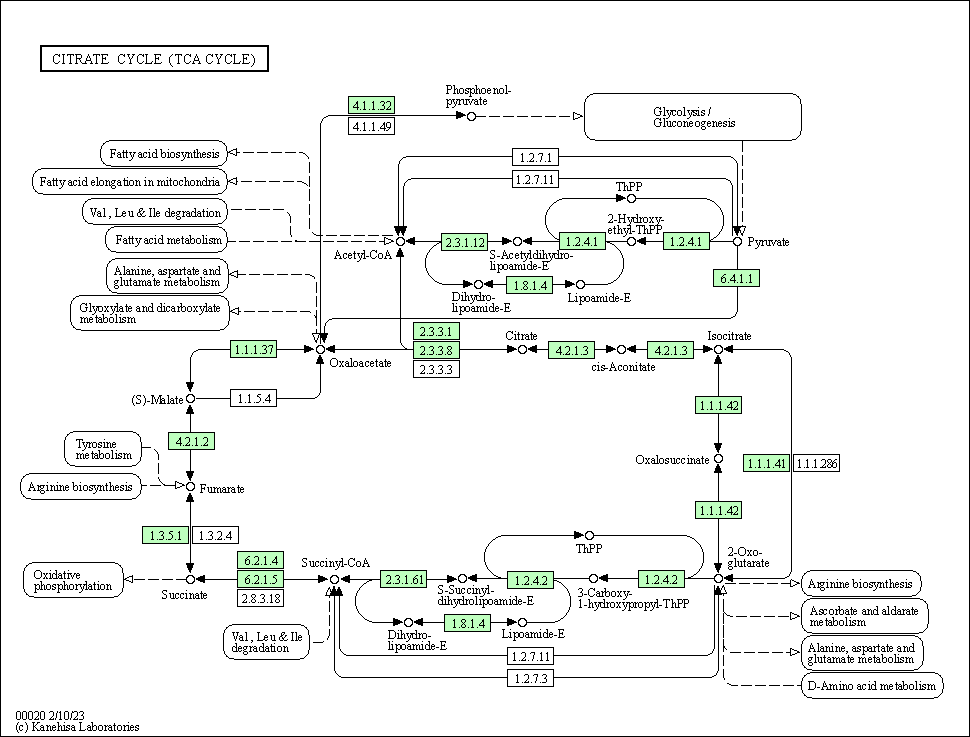
|
|||||||
| Metabolic pathways | hsa01100 |
Pathway Map 
|
|||||||
| Carbon metabolism | hsa01200 |
Pathway Map 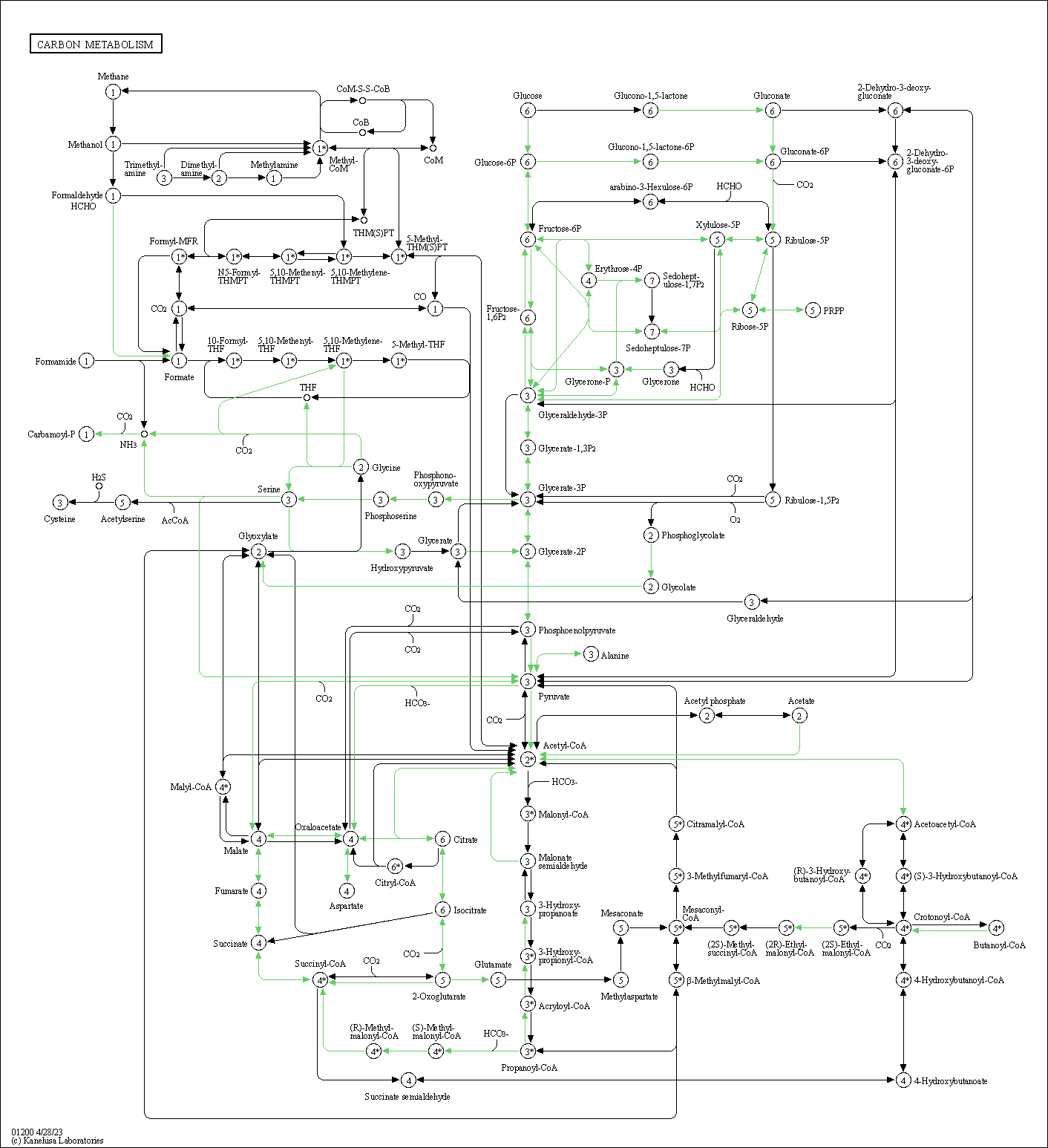
|
|||||||
| 2-Oxocarboxylic acid metabolism | hsa01210 |
Pathway Map 
|
|||||||
| Biosynthesis of amino acids | hsa01230 |
Pathway Map 
|
|||||||
| Glutathione metabolism | hsa00480 |
Pathway Map 
|
|||||||
| Peroxisome | hsa04146 |
Pathway Map 
|
|||||||
| 3D Structure |
|
||||||||
Full List of Virus RNA Interacting with This Protien
| Virus Name | Binding Region | RNA Info | Detection Method | Infection Cell | Cell ID | Cell Originated Tissue | Infection Time | Interaction Score 1 | Interaction Score 2 |
|---|---|---|---|---|---|---|---|---|---|
| Dengue virus | 5'UTR - 3'UTR | RNA Info | Comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS) | HuH-7.5.1 Cells (Human hepatocellular carcinoma cell) | . | Liver | 48 h | . | . |
| Zika virus | 5'UTR - 3'UTR | RNA Info | Comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS) | HuH-7.5.1 Cells (Human hepatocellular carcinoma cell) | . | Liver | 48 h | . | . |
| Encephalomyocarditis virus | 5'UTR - 3'UTR | RNA Info | Comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS) | HuH-7.5 Cells (Human hepatocellular carcinoma cell) | . | Liver | . | Z-score = -4.50546536 | FZR = 0.003225286 |
| Influenza A virus | 5'UTR - 3'UTR | RNA Info | Comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS) | HuH-7.5 Cells (Human hepatocellular carcinoma cell) | . | Liver | . | Z-score = 3.714627877 | FZR = 0.168907441 |
| Severe acute respiratory syndrome coronavirus 2 (strain hCoV-19/USA/WA1/2020) | Not Specified Virus Region | RNA Info | ChIRP-MS | Huh7.5 cells (Hepatocyte derived cellular carcinoma cell) | . | Liver | 48 h | FDR <= 0.05 | . |
| Severe acute respiratory syndrome coronavirus 2 (strain hCoV-19/Not Specified Virus Strain) | Not Specified Virus Region | RNA Info | RNAantisensepurificationandquantitativemassspectrometry(RAP-MS); Tandemmasstag(TMT)labelling; liquidchromatographycoupledwithtandemmassspectrometry(LC-MS/MS); Westernblot | Huh7 cells (liver carcinoma celll ine) | . | Liver | 24h | log2 fold change = 2.99294E+14 | . |
| Dengue virus 2 (strain 16681) | 5'UTR - 3'UTR | RNA Info | Comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS) | HuH-7.5.1 Cells (Human hepatocellular carcinoma cell) | . | Liver | 48 h | Saint score = 1 | MIST = 1.505 |
| Dengue virus 2 (strain 16681) | 5'UTR - 3'UTR | RNA Info | Comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS) | HuH-7.5.1 Cells (Human hepatocellular carcinoma cell) | . | Liver | 48 h | Saint score = 1 | MIST = 0.000 |
Potential Drug(s) that Targets This Protein
Protein Sequence Information
|
MSKKISGGSVVEMQGDEMTRIIWELIKEKLIFPYVELDLHSYDLGIENRDATNDQVTKDAAEAIKKHNVGVKCATITPDEKRVEEFKLKQMWKSPNGTIRNILGGTVFREAIICKNIPRLVSGWVKPIIIGRHAYGDQYRATDFVVPGPGKVEITYTPSDGTQKVTYLVHNFEEGGGVAMGMYNQDKSIEDFAHSSFQMALSKGWPLYLSTKNTILKKYDGRFKDIFQEIYDKQYKSQFEAQKIWYEHRLIDDMVAQAMKSEGGFIWACKNYDGDVQSDSVAQGYGSLGMMTSVLVCPDGKTVEAEAAHGTVTRHYRMYQKGQETSTNPIASIFAWTRGLAHRAKLDNNKELAFFANALEEVSIETIEAGFMTKDLAACIKGLPNVQRSDYLNTFEFMDKLGENLKIKLAQAKL
Click to Show/Hide
|
