Host Protein General Information
| Protein Name |
Cell division control protein 42 homolog
|
Gene Name |
CDC42
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| Host Species |
Homo sapiens
|
Uniprot Entry Name |
CDC42_HUMAN
|
||||||
| Protein Families |
Small GTPase superfamily, Rho family, CDC42 subfamily
|
||||||||
| EC Number |
3.6.5.2
|
||||||||
| Subcellular Location |
Cell membrane
|
||||||||
| External Link | |||||||||
| NCBI Gene ID | |||||||||
| Uniprot ID | |||||||||
| Ensembl ID | |||||||||
| HGNC ID | |||||||||
| Related KEGG Pathway | |||||||||
| Chemokine signaling pathway | hsa04062 |
Pathway Map 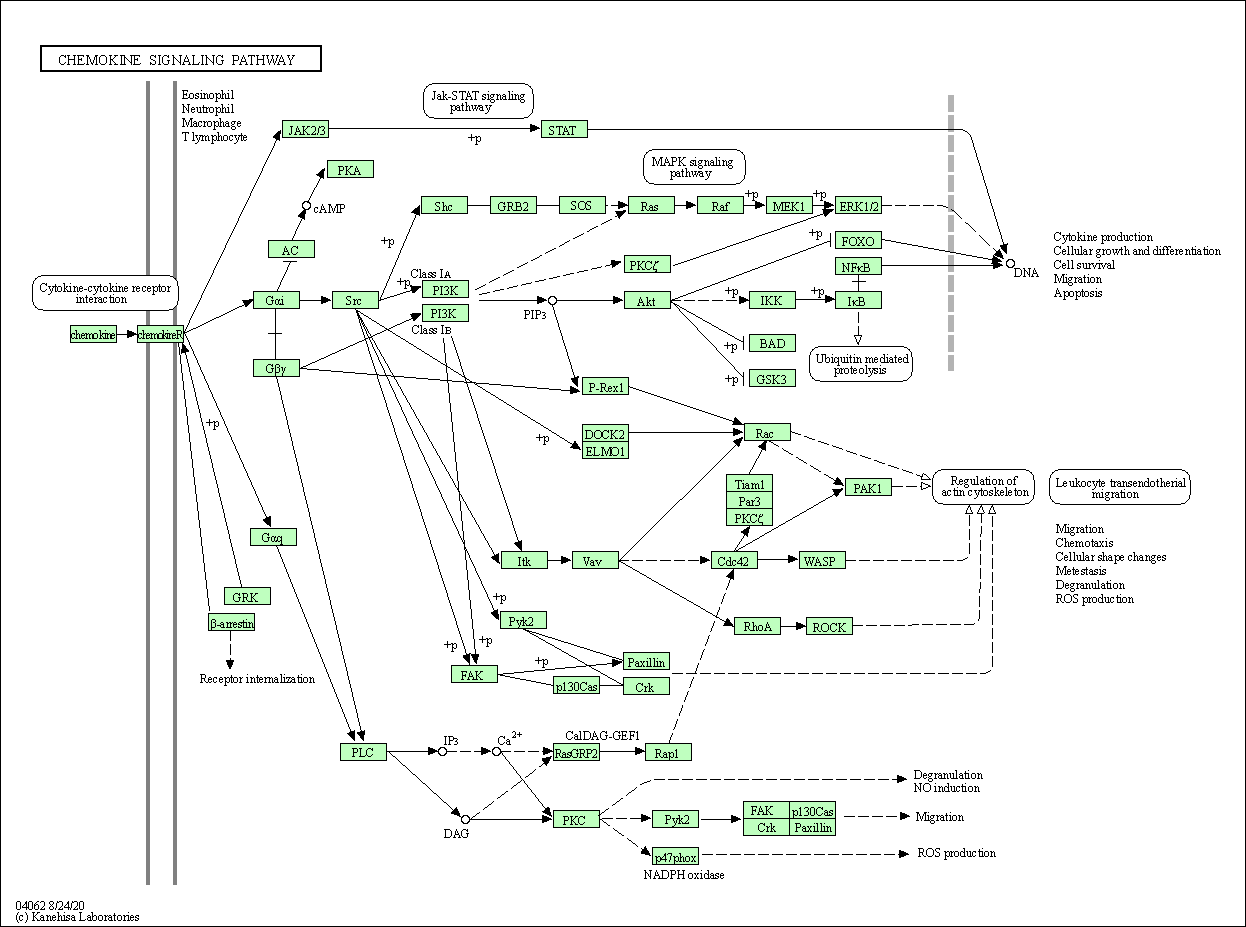
|
|||||||
| T cell receptor signaling pathway | hsa04660 |
Pathway Map 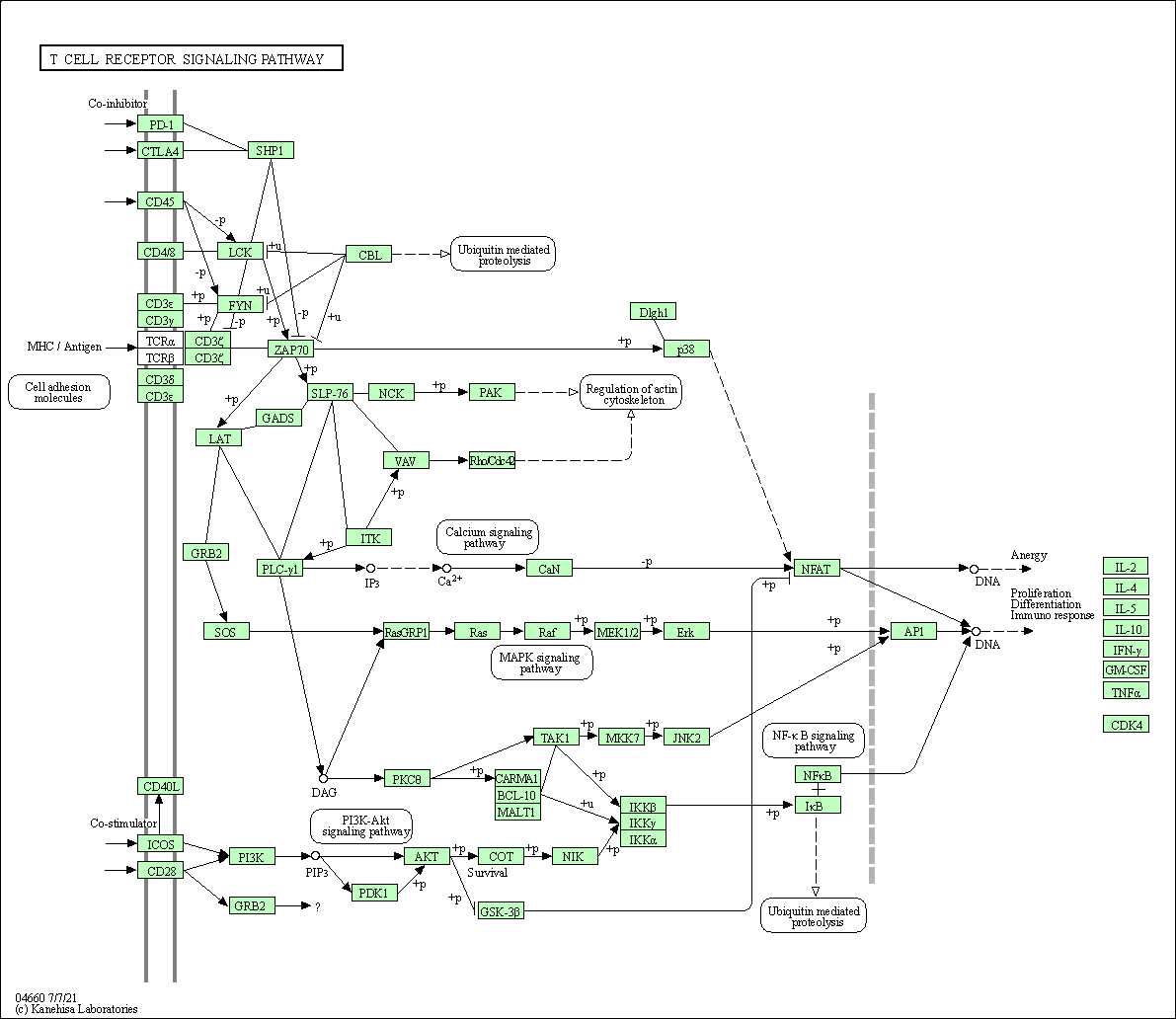
|
|||||||
| Fc gamma R-mediated phagocytosis | hsa04666 |
Pathway Map 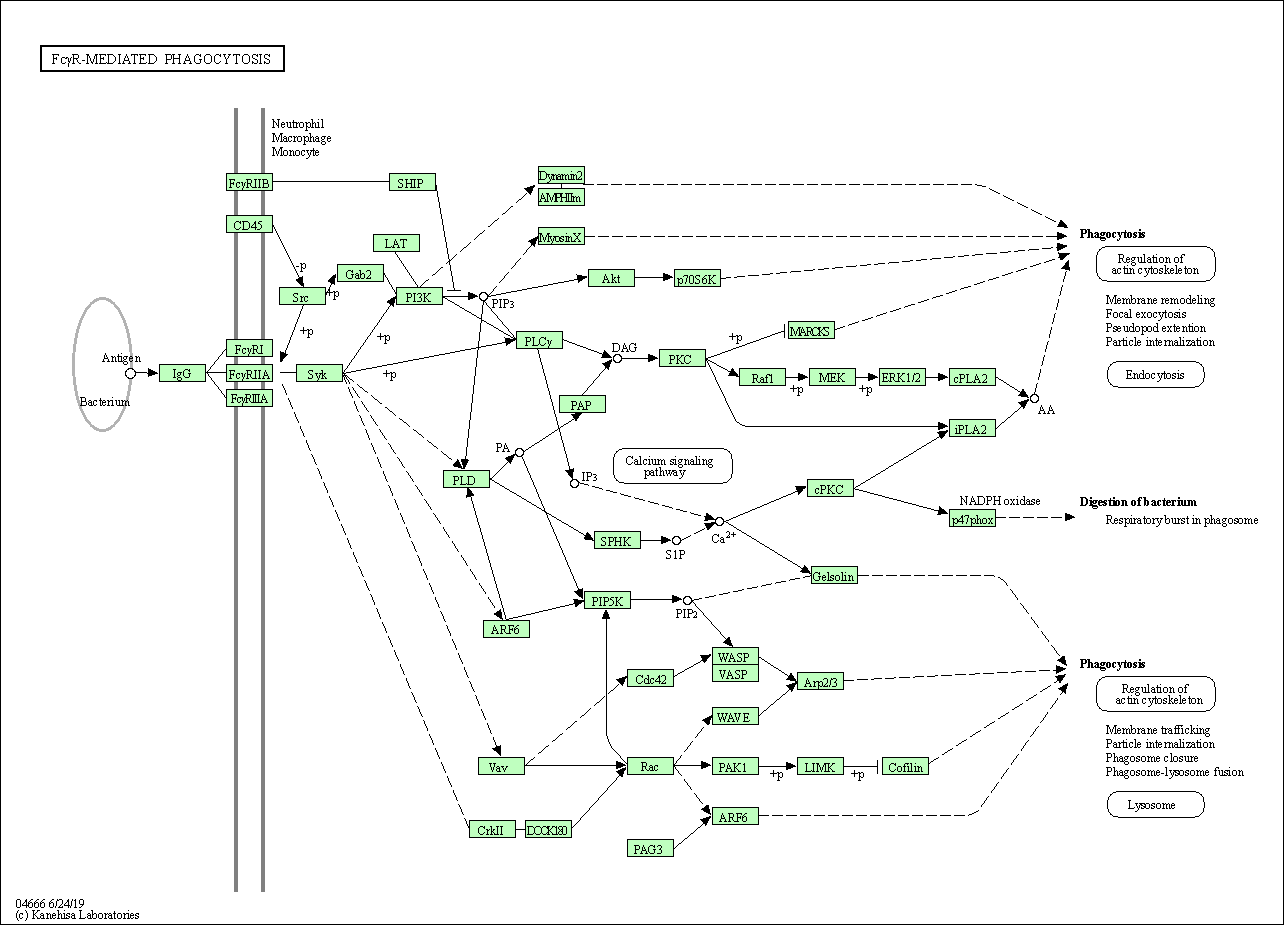
|
|||||||
| Leukocyte transendothelial migration | hsa04670 |
Pathway Map 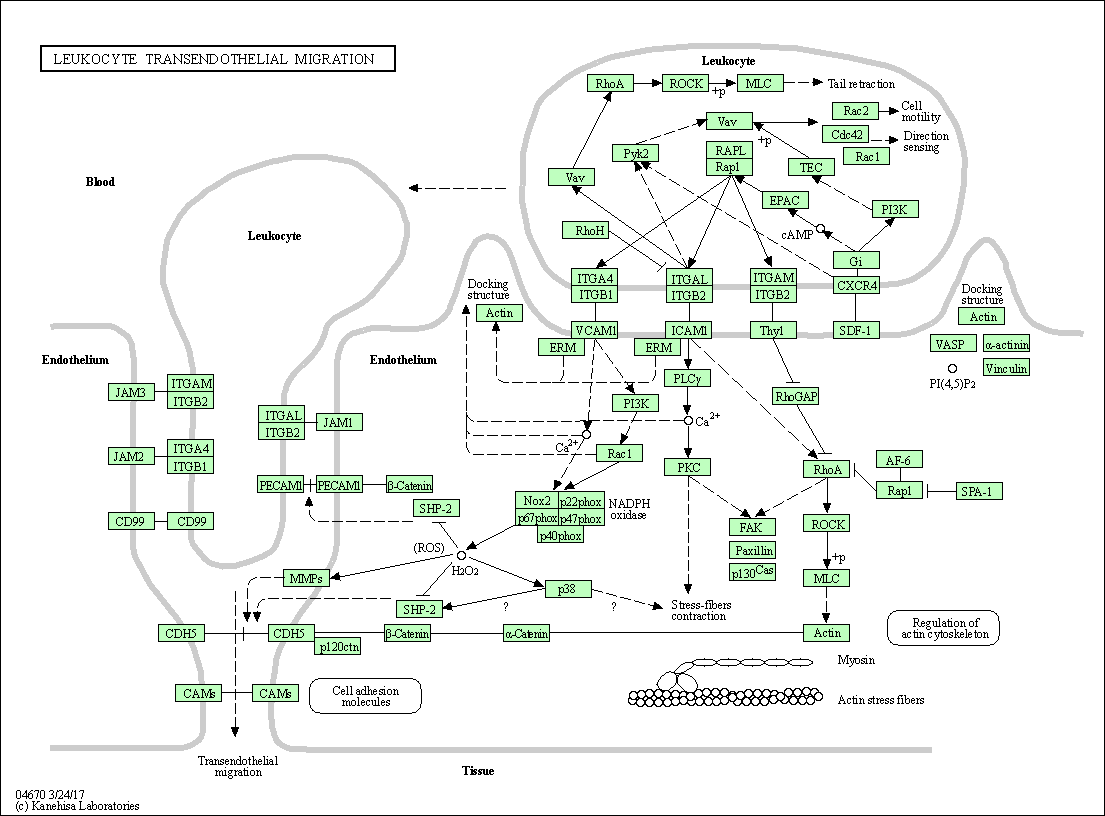
|
|||||||
| Bacterial invasion of epithelial cells | hsa05100 |
Pathway Map 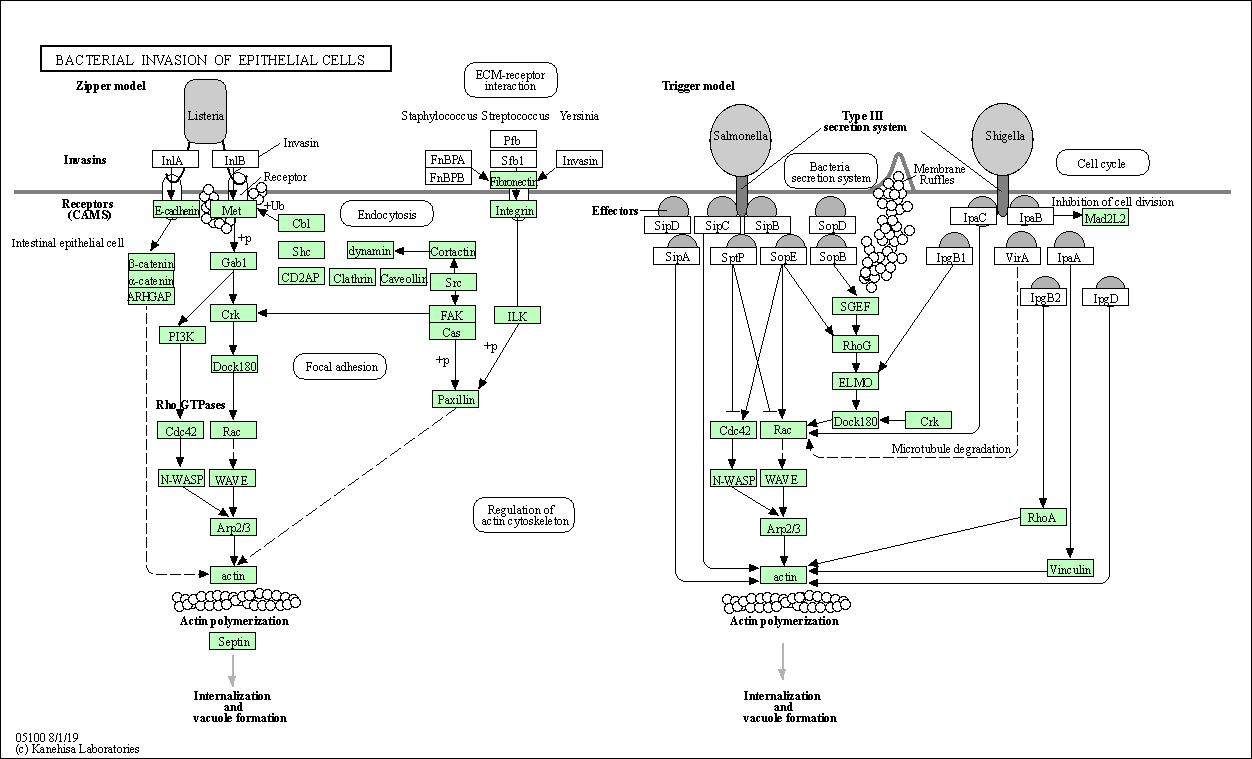
|
|||||||
| Epithelial cell signaling in Helicobacter pylori infection | hsa05120 |
Pathway Map 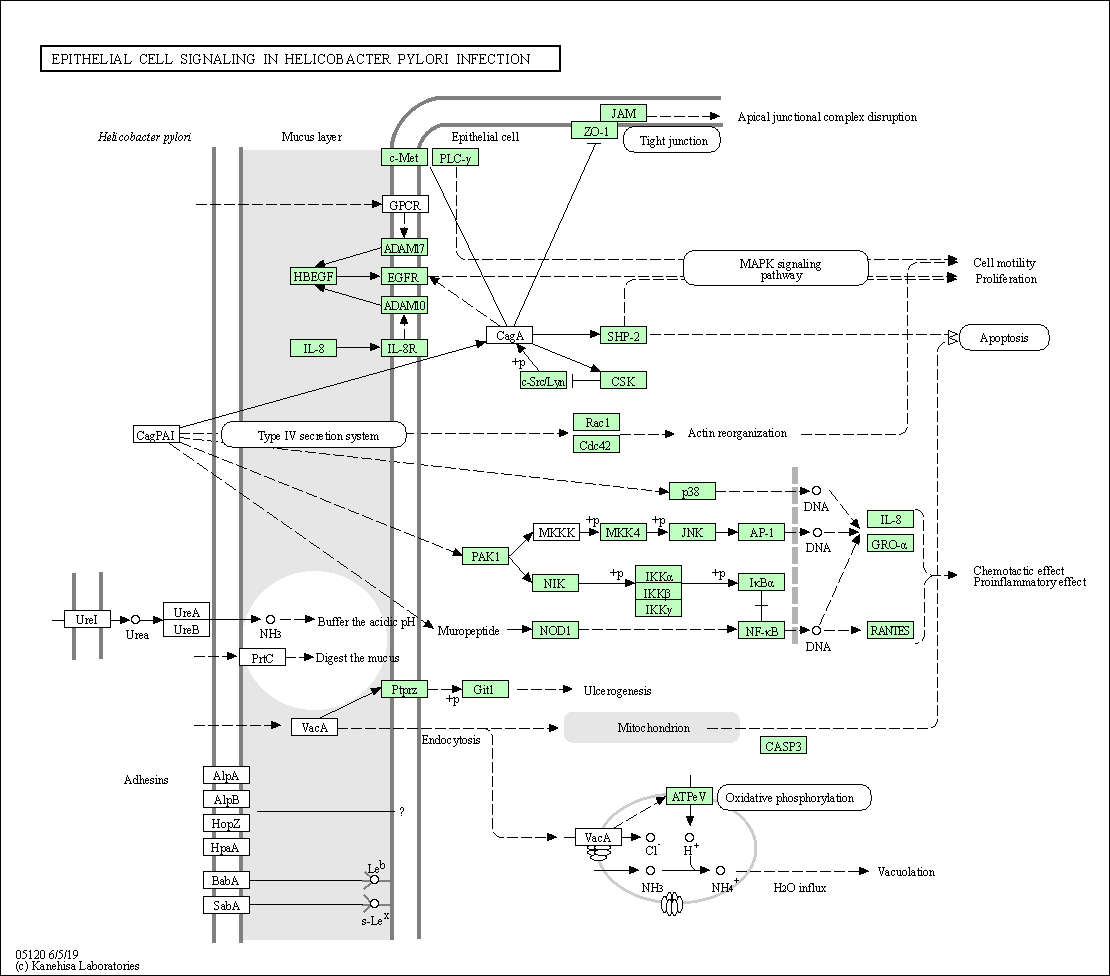
|
|||||||
| Pathogenic Escherichia coli infection | hsa05130 |
Pathway Map 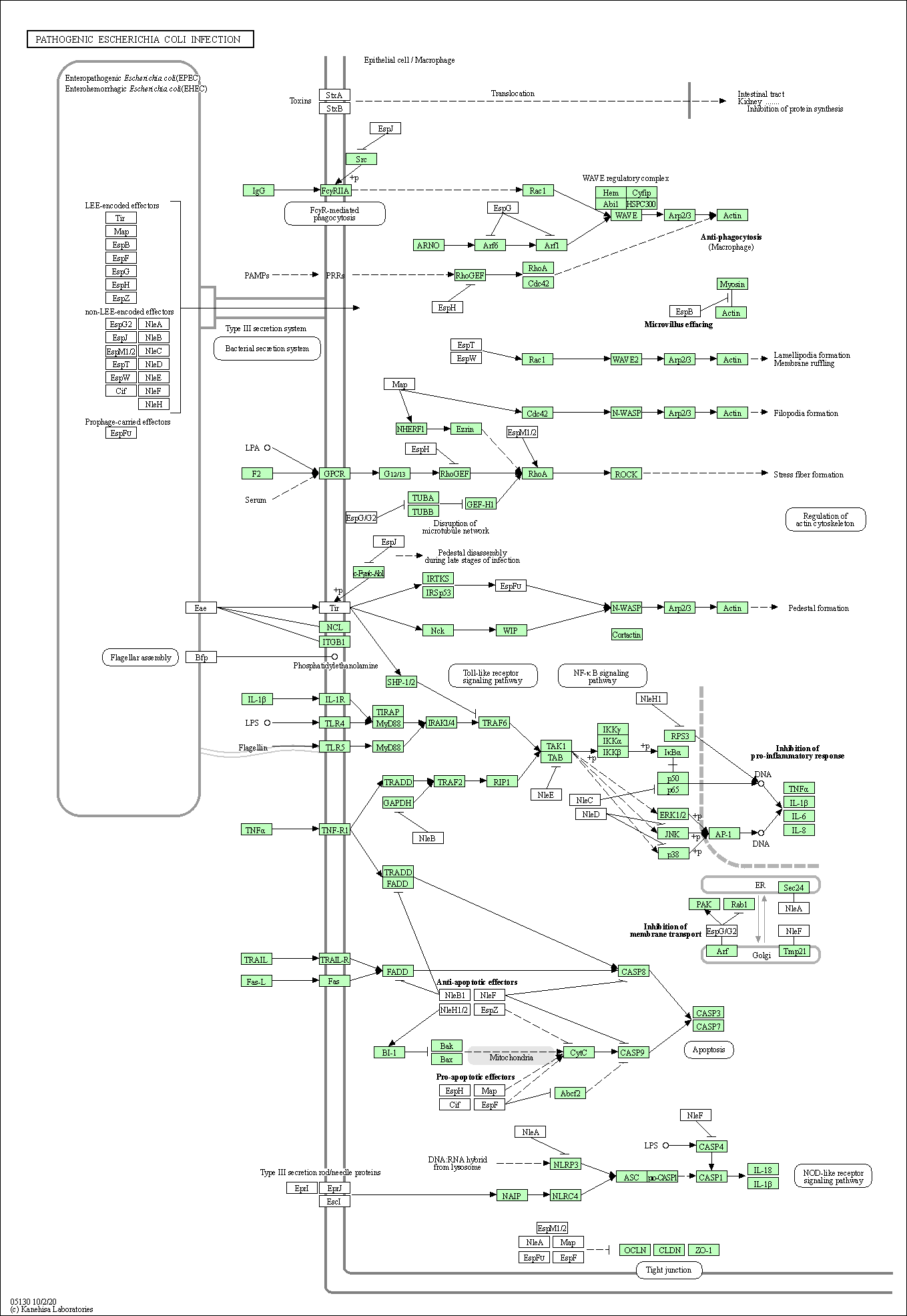
|
|||||||
| Shigellosis | hsa05131 |
Pathway Map 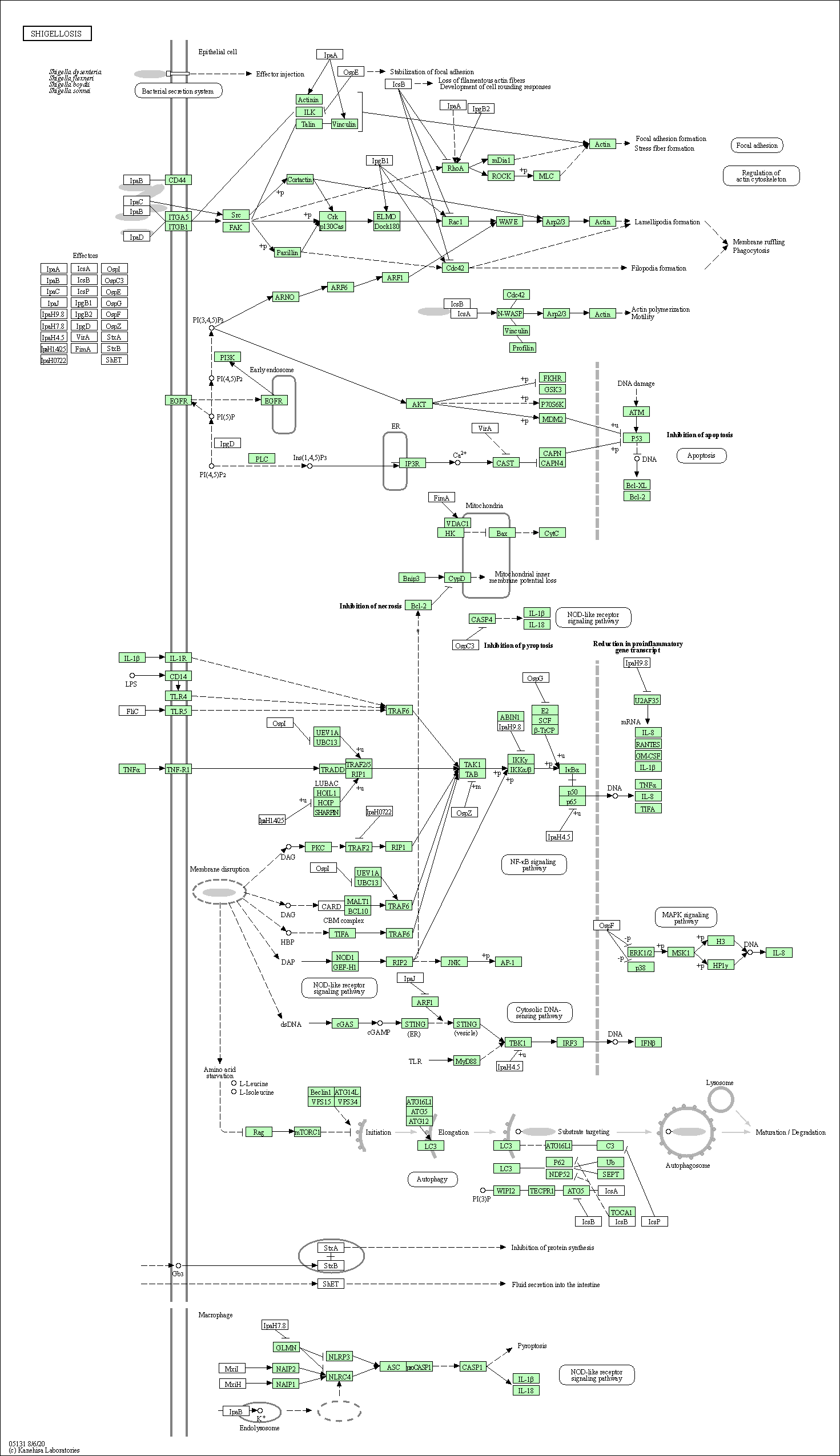
|
|||||||
| Salmonella infection | hsa05132 |
Pathway Map 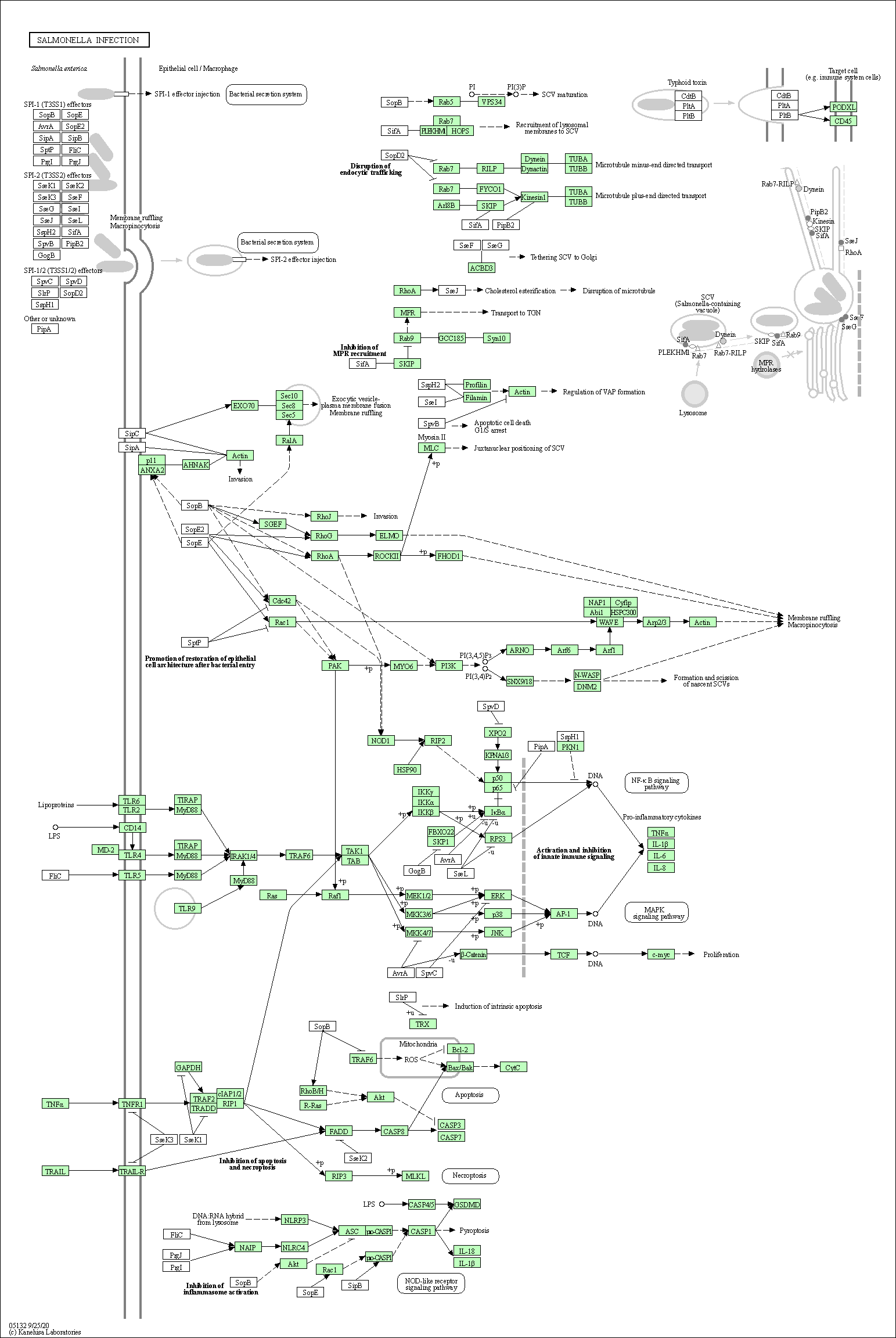
|
|||||||
| Yersinia infection | hsa05135 |
Pathway Map 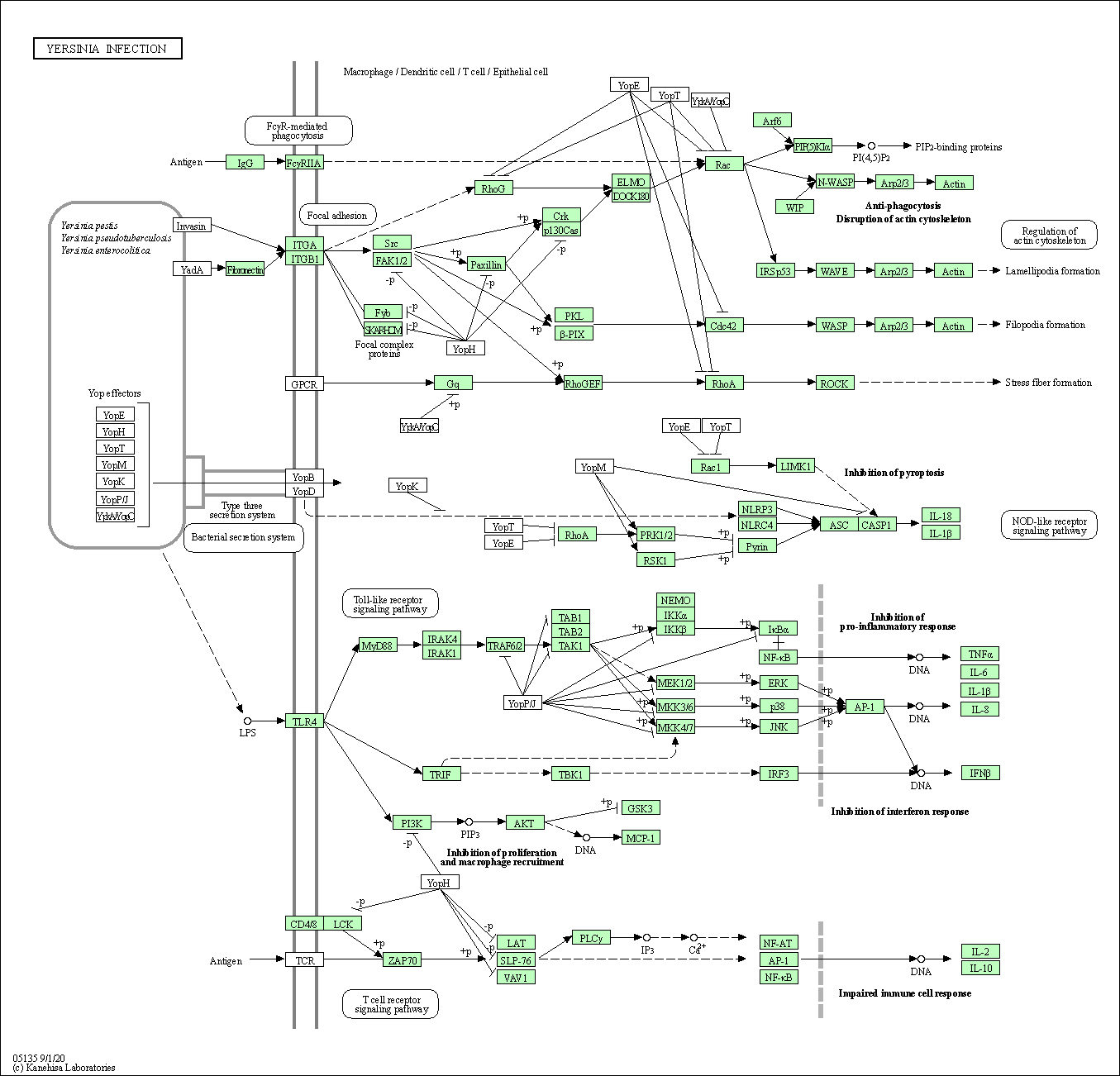
|
|||||||
| Human papillomavirus infection | hsa05165 |
Pathway Map 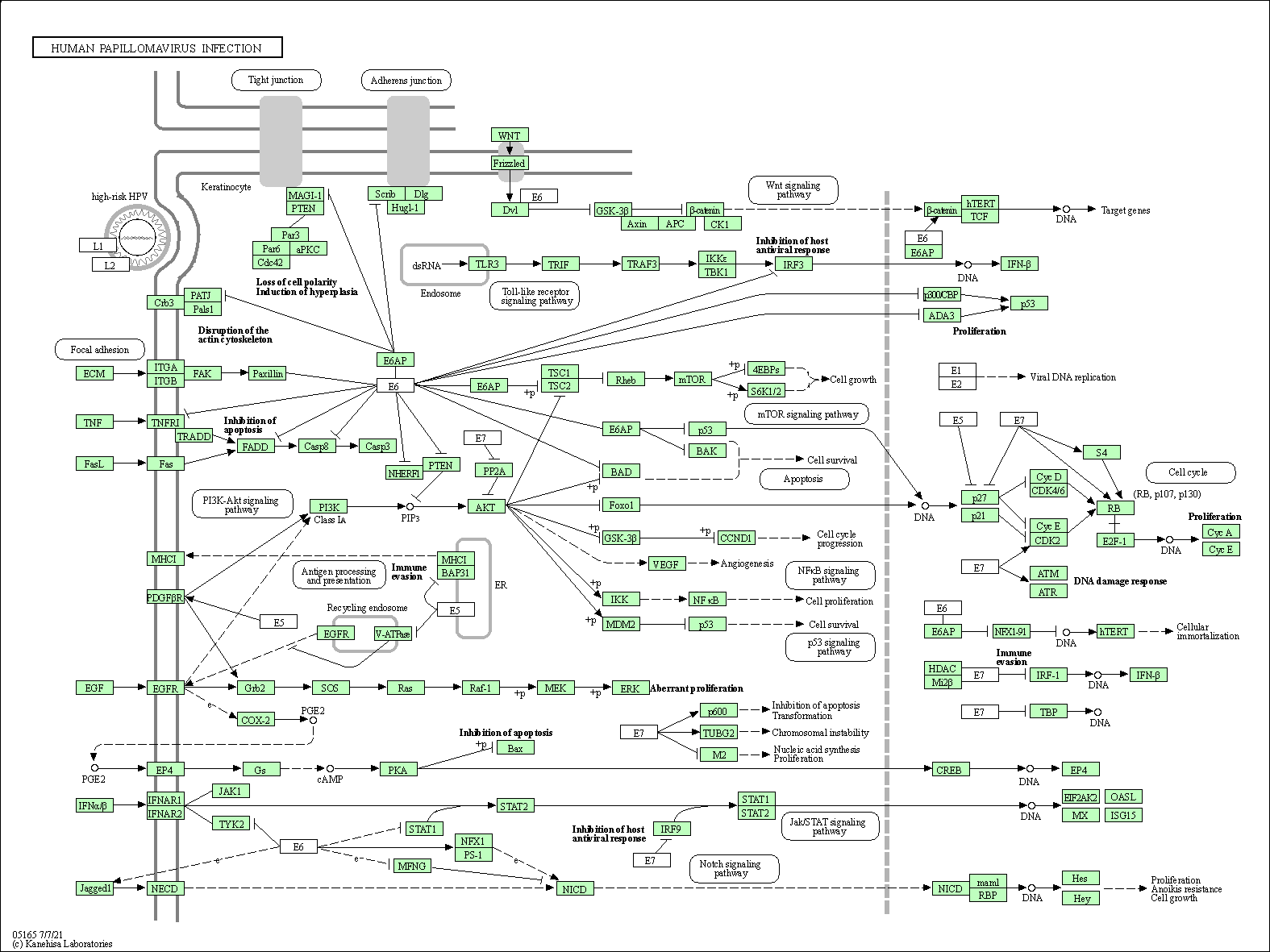
|
|||||||
| MAPK signaling pathway | hsa04010 |
Pathway Map 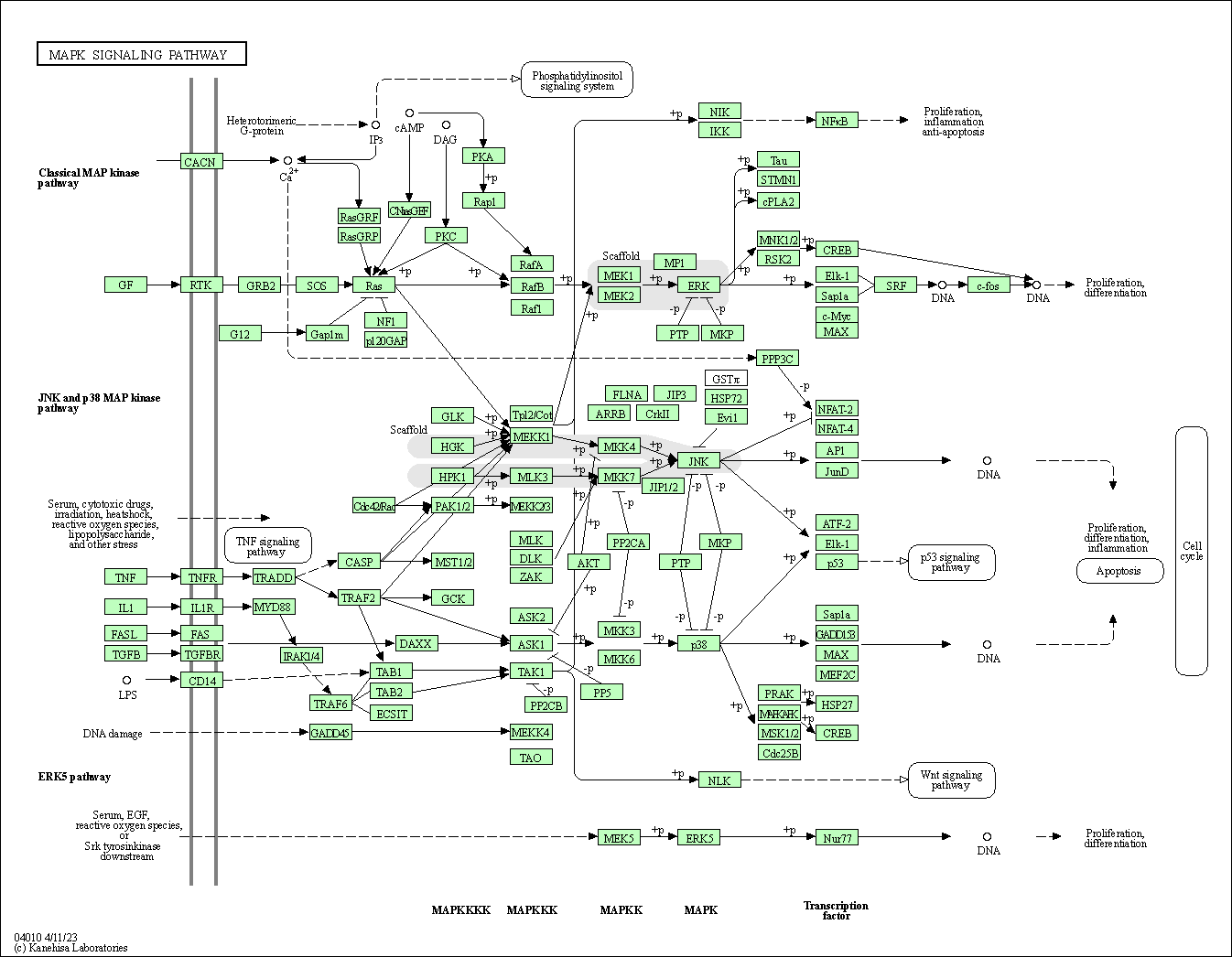
|
|||||||
| Ras signaling pathway | hsa04014 |
Pathway Map 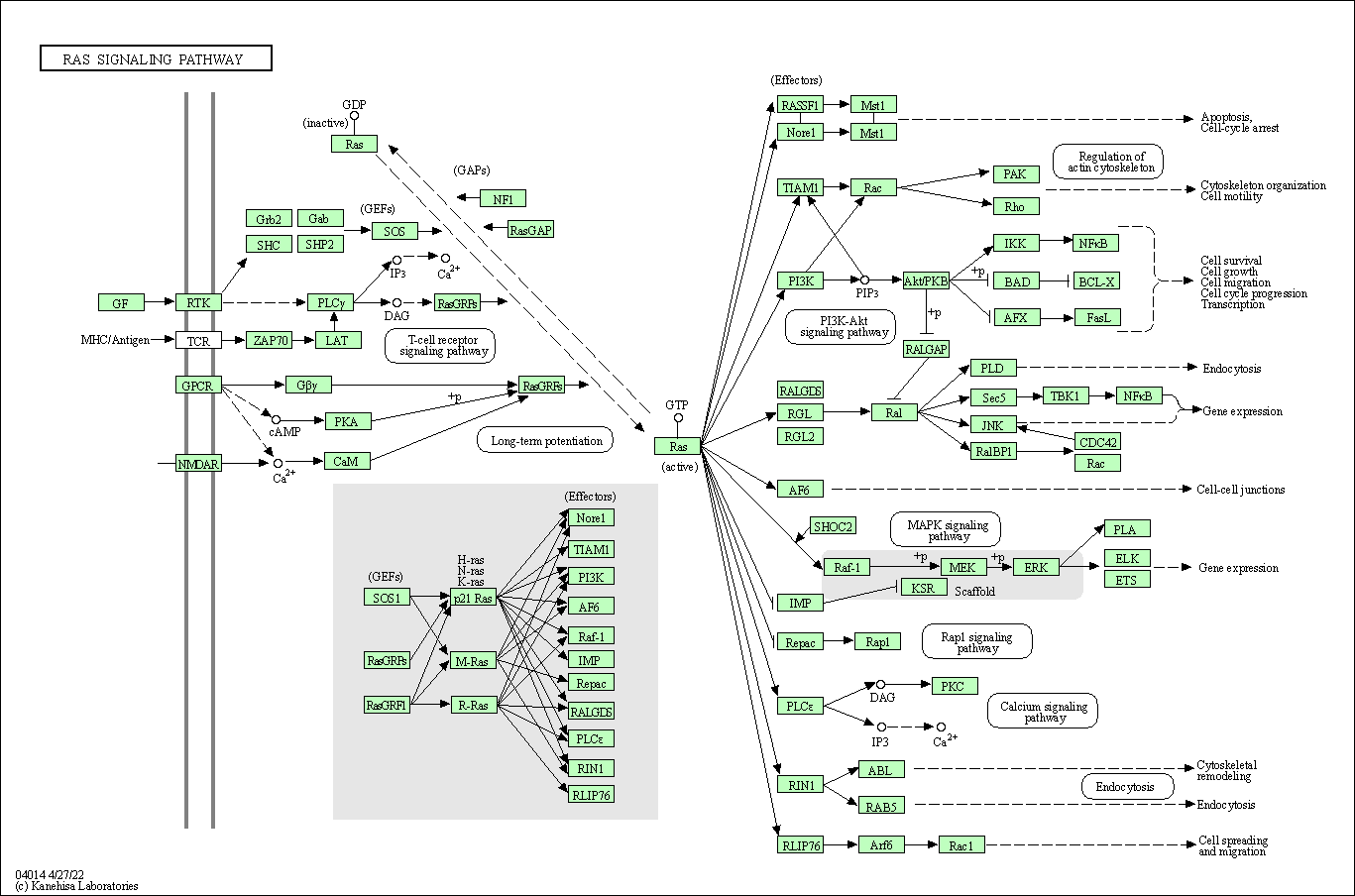
|
|||||||
| Rap1 signaling pathway | hsa04015 |
Pathway Map 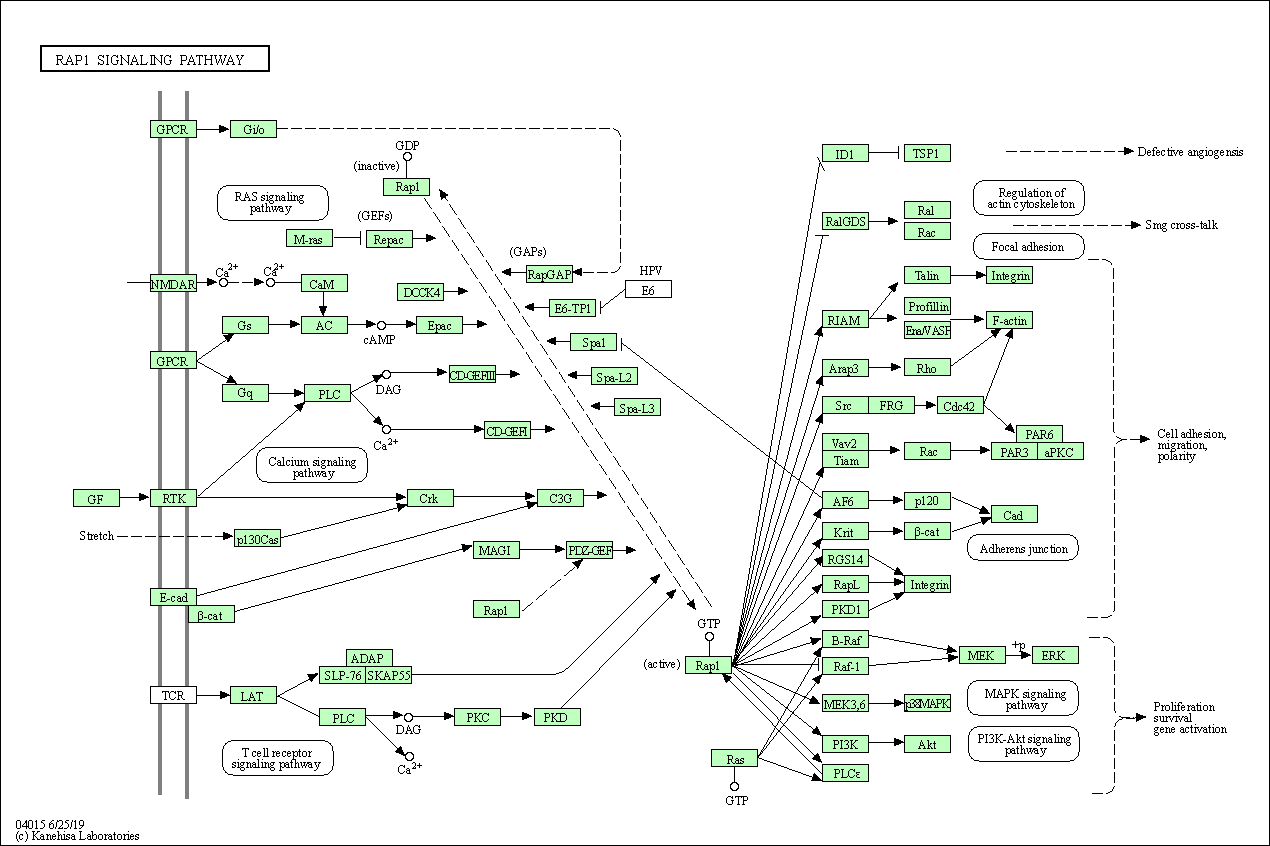
|
|||||||
| Endocytosis | hsa04144 |
Pathway Map 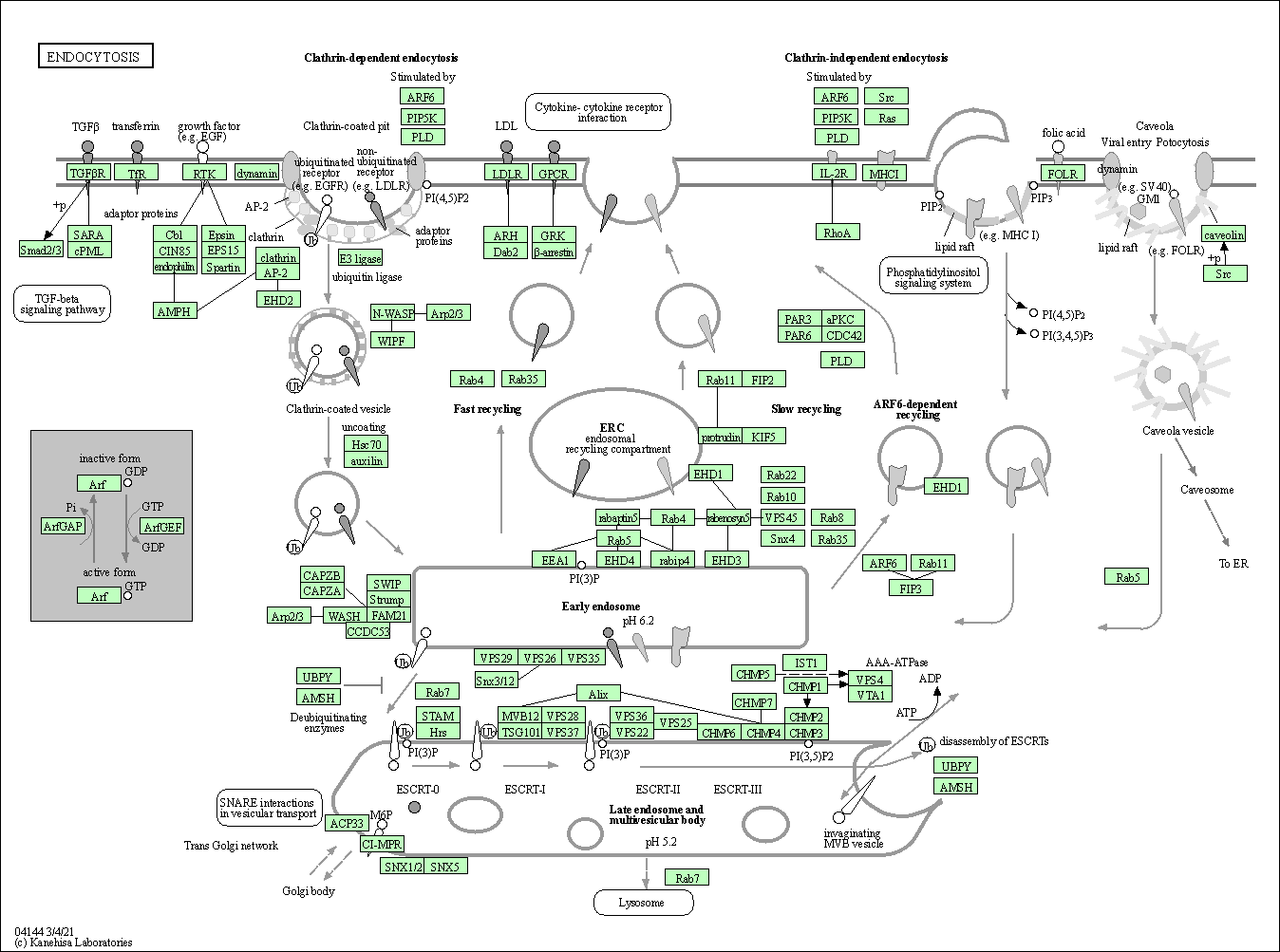
|
|||||||
| Axon guidance | hsa04360 |
Pathway Map 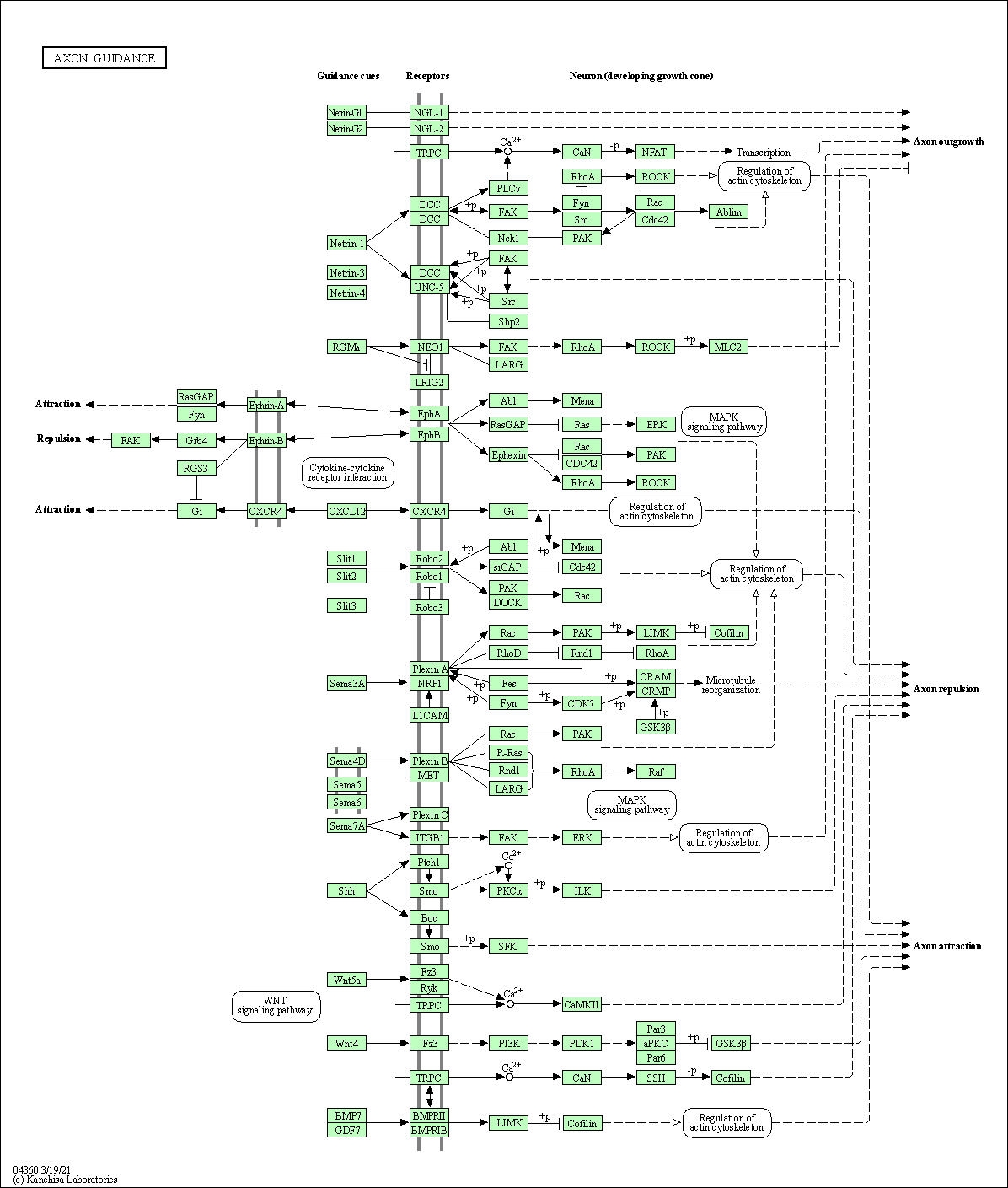
|
|||||||
| VEGF signaling pathway | hsa04370 |
Pathway Map 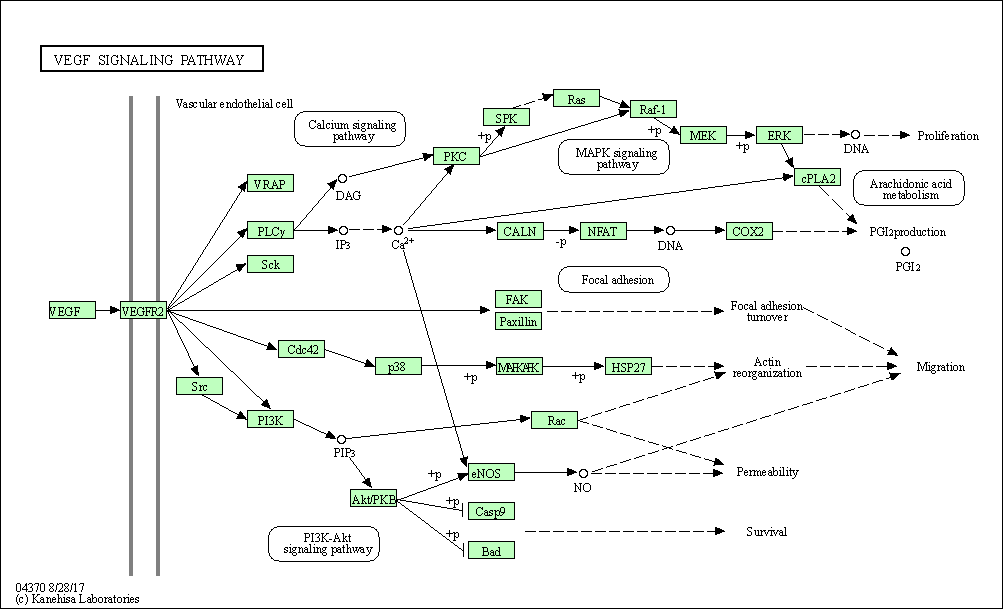
|
|||||||
| Focal adhesion | hsa04510 |
Pathway Map 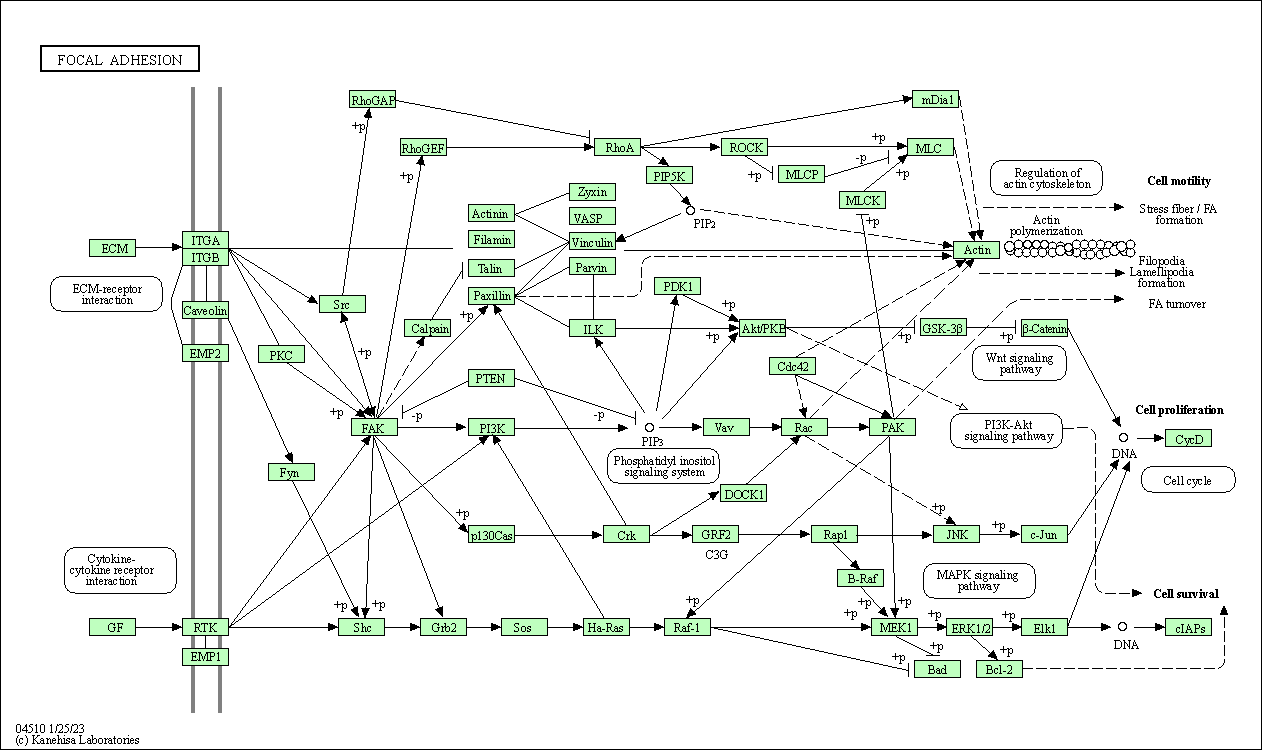
|
|||||||
| Adherens junction | hsa04520 |
Pathway Map 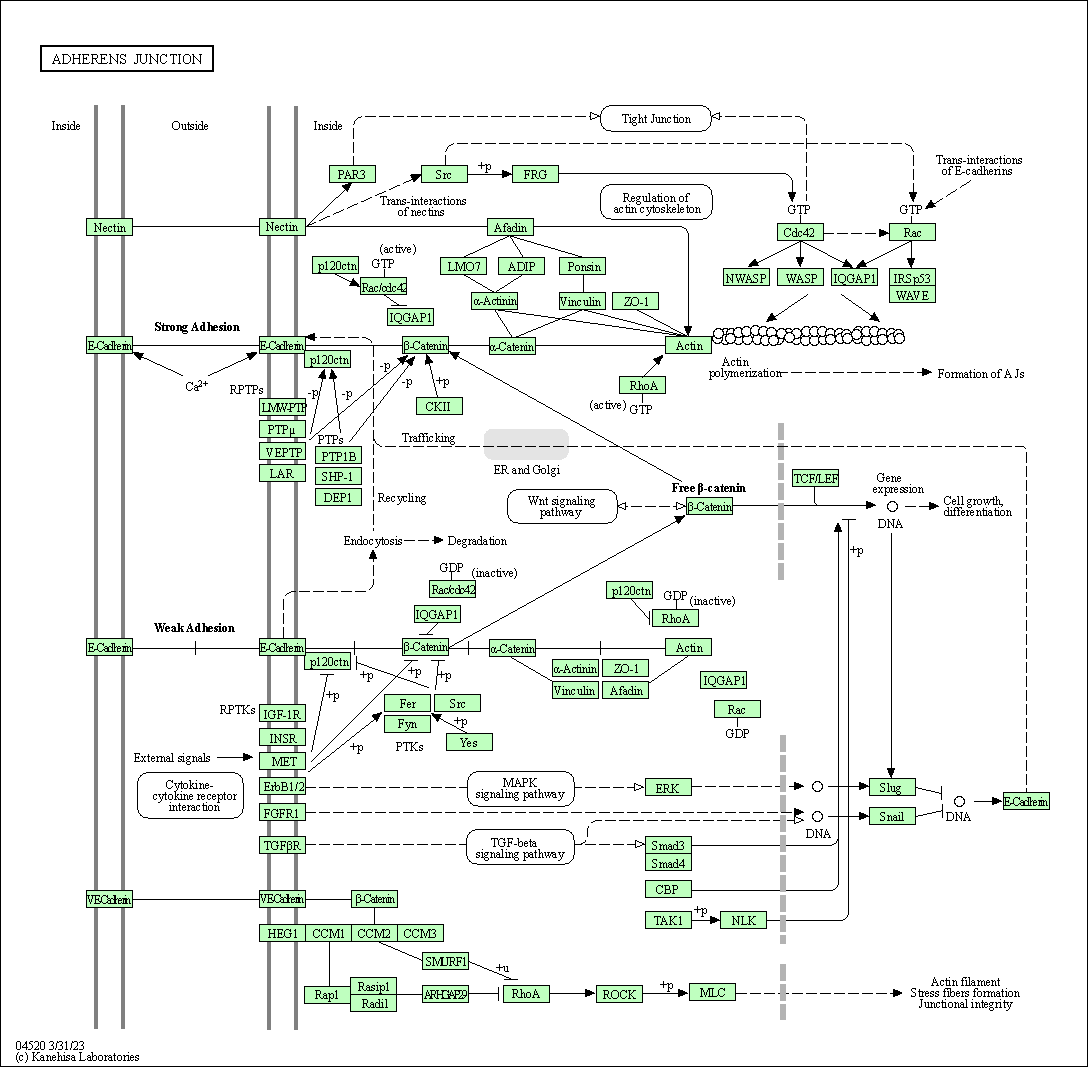
|
|||||||
| Tight junction | hsa04530 |
Pathway Map 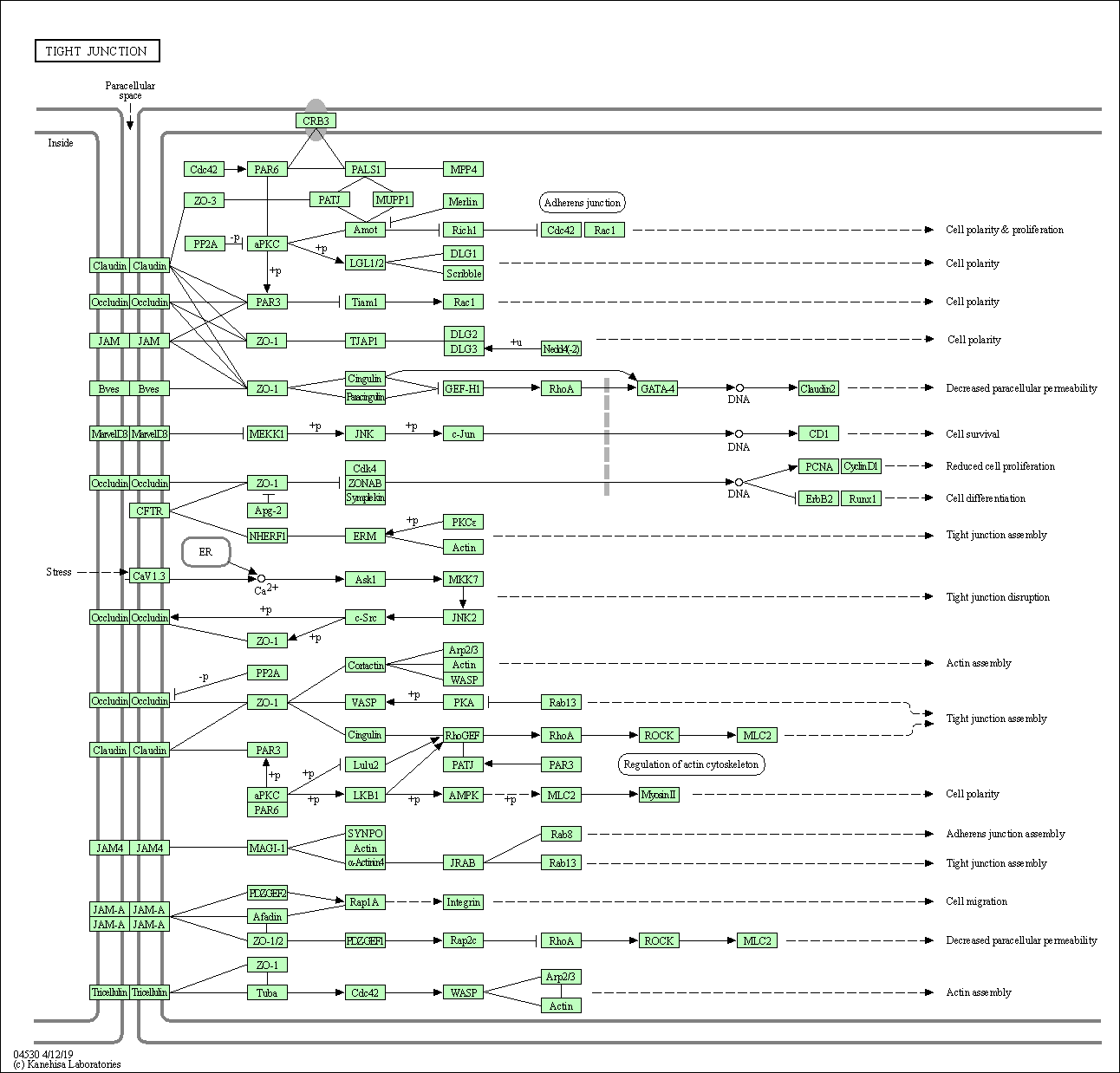
|
|||||||
| Neurotrophin signaling pathway | hsa04722 |
Pathway Map 
|
|||||||
| Regulation of actin cytoskeleton | hsa04810 |
Pathway Map 
|
|||||||
| GnRH signaling pathway | hsa04912 |
Pathway Map 
|
|||||||
| Non-alcoholic fatty liver disease | hsa04932 |
Pathway Map 
|
|||||||
| AGE-RAGE signaling pathway in diabetic complications | hsa04933 |
Pathway Map 
|
|||||||
| Pathways in cancer | hsa05200 |
Pathway Map 
|
|||||||
| Viral carcinogenesis | hsa05203 |
Pathway Map 
|
|||||||
| Proteoglycans in cancer | hsa05205 |
Pathway Map 
|
|||||||
| Renal cell carcinoma | hsa05211 |
Pathway Map 
|
|||||||
| Pancreatic cancer | hsa05212 |
Pathway Map 
|
|||||||
| Lipid and atherosclerosis | hsa05417 |
Pathway Map 
|
|||||||
| 3D Structure |
|
||||||||
Full List of Virus RNA Interacting with This Protien
| Virus Name | Binding Region | RNA Info | Detection Method | Infection Cell | Cell ID | Cell Originated Tissue | Infection Time | Interaction Score 1 | Interaction Score 2 |
|---|---|---|---|---|---|---|---|---|---|
| Encephalomyocarditis virus | 5'UTR - 3'UTR | RNA Info | Comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS) | HuH-7.5 Cells (Human hepatocellular carcinoma cell) | . | Liver | . | Z-score = -6.731783481 | FZR = 1.16144E-08 |
| Influenza A virus | 5'UTR - 3'UTR | RNA Info | Comprehensive identification of RNA-binding proteins by mass spectrometry (ChIRP-MS) | HuH-7.5 Cells (Human hepatocellular carcinoma cell) | . | Liver | . | Z-score = -0.388707323 | FZR = 1 |
| Influenza A virus (strain California/7/2004 (H3N2)) | 5'UTR - 3'UTR | RNA Info | VIRal Cross-Linking And Solid-phase Purification (VIR-CLASP) | BHK-21 Cells (Small hamster kidney fibroblast) | . | Kidney | 1 h | P = 0.000923885592436679 | . |
| Chikungunya virus (strain 181/25) | 5'UTR - 3'UTR | RNA Info | VIRal Cross-Linking And Solid-phase Purification (VIR-CLASP) | BHK-21 Cells (Small hamster kidney fibroblast) | . | Kidney | 0.2 h | P = 6.57776343267714e-05 | . |
| Chikungunya virus (strain 181/25) | 5'UTR - 3'UTR | RNA Info | VIRal Cross-Linking And Solid-phase Purification (VIR-CLASP) | BHK-21 Cells (Small hamster kidney fibroblast) | . | Kidney | 1 h | P = 3.23924127896155e-05 | . |
| Chikungunya virus (strain 181/25) | 5'UTR - 3'UTR | RNA Info | VIRal Cross-Linking And Solid-phase Purification (VIR-CLASP) | BHK-21 Cells (Small hamster kidney fibroblast) | . | Kidney | 3 h | P = 0.000134687073250936 | . |
Protein Sequence Information
|
MQTIKCVVVGDGAVGKTCLLISYTTNKFPSEYVPTVFDNYAVTVMIGGEPYTLGLFDTAGQEDYDRLRPLSYPQTDVFLVCFSVVSPSSFENVKEKWVPEITHHCPKTPFLLVGTQIDLRDDPSTIEKLAKNKQKPITPETAEKLARDLKAVKYVECSALTQKGLKNVFDEAILAALEPPEPKKSRRCVLL
Click to Show/Hide
|
